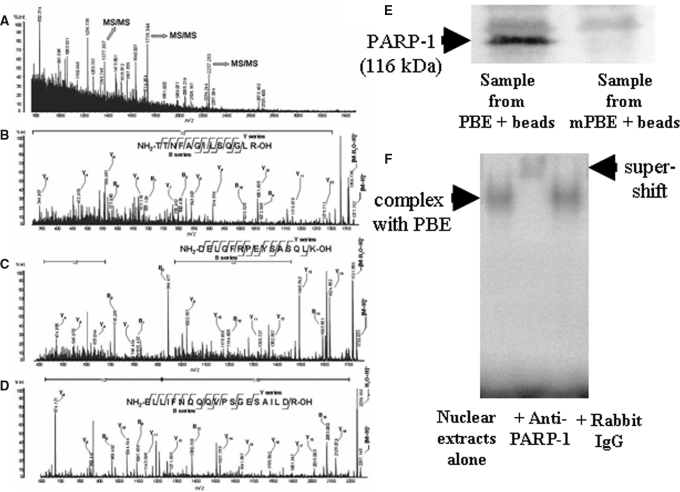
Figure 5.
Representative MALDI-TOF MS spectra in PMF analysis (A) and de novo sequence analyses (B through D). MS/MS spectra of each parent peptide ion m/z 1377.807, 1739.944, and 2257.283 in A are B, C, and D, respectively. Bn and Yn show generic description of potential ions that are formed by fragmentation of a parent peptide. The Bn and Yn ions are fragments that retain the charge at the amino end (N-terminus) and carboxy end (C-terminus) of the peptide molecules, respectively. Location and N-terminus–or C-terminus–derived direction of the fragment ions that reflect the peptide bonds in the database search are shown with amino acid sequence of the parent peptides. (E) Nuclear protein purified using the biotin-streptavidin DNA affinity technique with the identified putative DNA binding sequence (PBE) was targeted for immunoblotting, which revealed that the trans-factor protein was a 116 kD, PARP-1 protein. mPBE, mutated PBE. (F) Anti–PARP-1 antibody caused a supershift of the shifted band of the −455- to −434-bp CCN2 promoter fragment (PBE) bound to the nuclear protein extracts in the EMSA analysis, but application of rabbit IgG was without effects, indicating that PARP-1 protein interacted with the PBE. The blot is representative of four independent experiments.