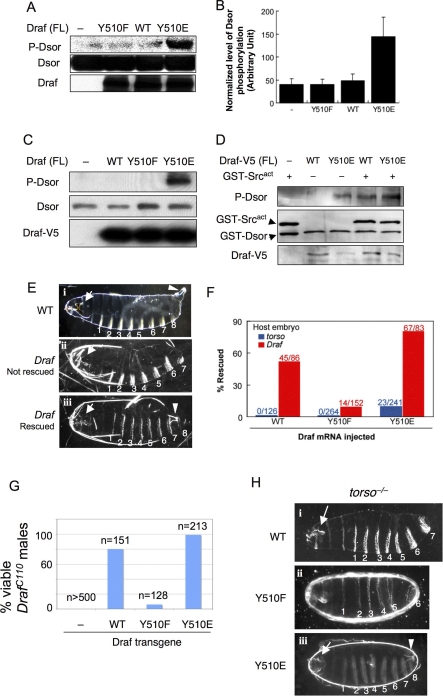
Figure 3. Acidic Substitution of Y510 Promotes and Phenylalanine Substitution Impairs Activation of Full-Length Draf In Vitro and In Vivo.
(A) DrafY510E (lane 4) exhibited dramatically higher activities by in vitro kinase assay using bacterially expressed full-length Draf variants and GST-Dsor1. Dsor1 phosphorylation was detected by anti-pMEK. WT, wild type.
(B) Quantification of results represented in (A).
(C) V5-tagged full-length Draf variants were transfected into S2 cells and were immunoprecipitated with anti-V5 and subjected to kinase assay using bacterially expressed Dsor1 as substrate. Note that only DrafY510E exhibited prominent kinase activity toward purified Dsor1, as detected by anti-pMEK (lane 4).
(D) V5-tagged full-length DrafWT or DrafY510E were transfected into S2 cells and were immunoprecipitated with anti-V5 and then subjected to kinase assay using bacterially expressed Dsor1 as substrate with or without purified GST-Src64Bact. Note that GST-Src64Bact stimulated the activity of DrafWT to that of DrafY510E (cf. lanes 3 and 4).
(E–H) Effects of Y510 substitutions on Draf activity in vivo. (E and F) Injection of mRNA into early stage embryos. (i) Cuticles of a wild-type embryo exhibiting normal head skeleton (arrow), eight ventral denticle belts (numbered), and the Filzkörper (arrowhead). (ii) Cuticles from a buffer-injected (control) Draf −/− embryo exhibit characteristic Draf null phenotypes: collapsed head skeletons (arrow) and loss of all posterior structures (eighth denticle belt and the Filzkörper). (iii) A Draf null embryo rescued by injecting DrafY510E mRNA. Note the restored eighth denticle belt and the Filzkörper (arrowhead). Due to the limited diffusion of injected mRNA or different threshold requirement, the anterior (head skeleton; arrow) defects were not rescued, which serves as an internal control. The phenotypes of rescued torso null embryos are identical to Draf null embryos (unpublished data).
(F) DrafY510E expressed from injected mRNA exhibited higher basal and inducible activity than DrafWT (p < 0.001) in vivo. DrafY510F had impaired inducible activity (p < 0.001). Basal Draf activity was measured by the percentage of rescue of posterior structures of torso null embryos by mRNA microinjection (shown in blue). DrafY510E rescued 9.5% of injected torso embryos, whereas no rescue was found for DrafY510F and DrafWT. Inducible Draf activity was measured by the percentage of rescue of posterior structures of Draf null embryos by mRNA microinjection (shown in red). DrafWT, DrafY510F, or DrafY510E rescued 52%, 9.2%, or 81% of Draf null embryos, respectively. The number of injected embryos scored is indicated. Chi-square tests were used to compare the differences, p-values are indicated.
(G and H) In vivo activities of Draf transgenes with different Y510 substitutions. (G) Virgin DrafC110/FM7 females were mated with males with a recombinant chromosome carrying hsp70-Gal4 and an indicated UAS-Draf transgene. Surviving F1 DrafC110/Y males were scored and normalized as percent viability relative to control crosses. Note that DrafC110/Y males were not viable in the absence of UAS-Draf transgenes, but were rescued to different degrees by different Draf transgenes. (H) Females of Nanos-Gal4; torsoXR1; UAS-Draf variants were mated to wild-type males, and the F1 progeny were examined for viability and cuticle patterns. Note that expressing DrafWT or DrafY510F did not rescue torsoXR1 phenotypes, whereas expressing DrafY510E fully rescued torsoXR1 female sterility (unpublished data) or restored the posterior cuticle structures.