Abstract
Single-chain insulin (SCI) analogs provide insight into the inter-relation of hormone structure, function, and dynamics. Although compatible with wild-type structure, short connecting segments (<3 residues) prevent induced fit upon receptor binding and so are essentially without biological activity. Substantial but incomplete activity can be regained with increasing linker length. Here, we describe the design, structure, and function of a single-chain insulin analog (SCI-57) containing a 6-residue linker (GGGPRR). Native receptor-binding affinity (130 ± 8% relative to the wild type) is achieved as hindrance by the linker is offset by favorable substitutions in the insulin moiety. The thermodynamic stability of SCI-57 is markedly increased (ΔΔGu = 0.7 ± 0.1 kcal/mol relative to the corresponding two-chain analog and 1.9 ± 0.1 kcal/mol relative to wild-type insulin). Analysis of inter-residue nuclear Overhauser effects demonstrates that a native-like fold is maintained in solution. Surprisingly, the glycine-rich connecting segment folds against the insulin moiety: its central Pro contacts ValA3 at the edge of the hydrophobic core, whereas the final Arg extends the A1-A8 α-helix. Comparison between SCI-57 and its parent two-chain analog reveals striking enhancement of multiple native-like nuclear Overhauser effects within the tethered protein. These contacts are consistent with wild-type crystal structures but are ordinarily attenuated in NMR spectra of two-chain analogs, presumably due to conformational fluctuations. Linker-specific damping of fluctuations provides evidence for the intrinsic flexibility of an insulin monomer. In addition to their biophysical interest, ultrastable SCIs may enhance the safety and efficacy of insulin replacement therapy in the developing world.
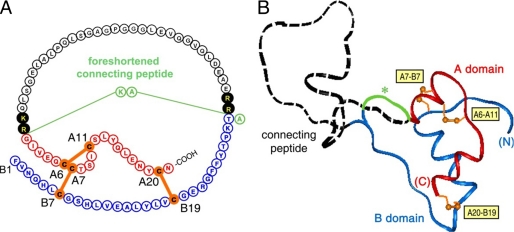
Insulin is a small globular protein containing two chains, A (21 residues) and B (30 residues). In pancreatic β-cells, the hormone is the product of a single-chain precursor, proinsulin, which contains a disordered connecting peptide between the B- and A-chains (Fig. 1) (1). Although proinsulin retains a native-like insulin moiety, its activity is >50-fold lower than that of insulin. Following excision of the connecting peptide (35 residues in human proinsulin) (Fig. 1A), the mature hormone is stored as Zn2+-stabilized hexamers within specialized secretory granules (1). The hexamers dissociate upon secretion into the portal circulation, enabling the hormone to function in the bloodstream as a zinc-free monomer. Although the structure of insulin has been extensively investigated by x-ray crystallography (2-5) and NMR spectroscopy (6-11), a variety of evidence suggests that the hormone undergoes a significant change in conformation upon receptor binding (for review, see Ref. 12). Foreshortened connecting segments (<3 residues) (illustrated in green in Fig. 1A) prevent induced fit upon receptor binding and so are essentially without biological activity (Fig. 1B, asterisk) (13-18).
FIGURE 1.
Structure of human proinsulin and inactive foreshortened analog. A, primary structure. The A-domain is shown in red, the B-domain in blue, and the connecting region in black. Disulfide bridges are shown in orange (A6-A11, A7-B7, and A20-B19). Filled circles highlight dibasic cleavage sites for prohormone convertases. PIP (relative activity of 0.14%) (18) contains a dipeptide linker as a foreshortened connecting segment (AK; light green) and a porcine insulin residue at B30 (Ala). B, structural model of human proinsulin depicting the insulin-like moiety and disordered connecting domain (dashed black line). The asterisk indicates the possible path of the foreshortened connecting segment (green). The coloring scheme is as described for A. Disulfide bonds are labeled in yellow boxes.
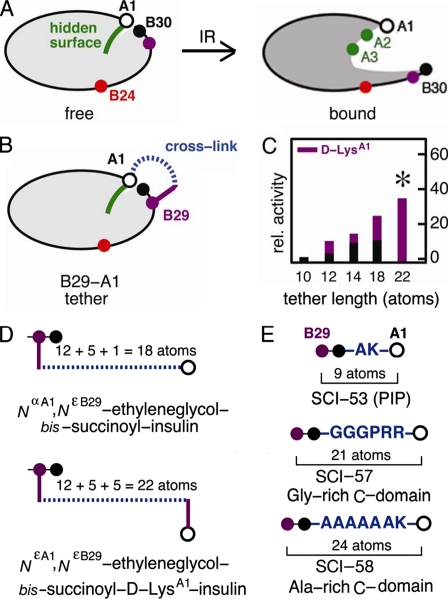
The detachment model envisages that, upon receptor binding, the C-terminal β-strand of the B-chain reorganizes to enable its binding between domains of the insulin receptor (IR),5 in turn exposing non-polar side chains of the A-chain as an otherwise “hidden” receptor-binding surface (6, 11, 14, 15, 19-23). Such induced fit may be regarded as a transition between closed and open conformational states (Fig. 2A, left and right panels). Whereas the side chains of IleA2 and ValA3 are buried in the free (closed) state, studies of structure-activity relationships suggest that they directly contact the IR (Fig. 2A, right panel, green circles) (11, 21, 23-25). Anomalies encountered in studies of substitutions at PheB24 (Fig. 2A, red circles) suggest that this site functions as a structural switch (6, 11, 20-22, 26, 27). This model rationalizes both the enhanced activities of nonstandard analogs containing d-amino acids at B24 (20, 26, 27) and the inactivity of markedly foreshortened proinsulin analogs (single-chain insulins (SCIs)) (13-17, 28). The closed conformation of insulin may reduce non-native aggregation and hence protect the β-cell from proteotoxicity (29).
FIGURE 2.
Model of induced fit and design of interchain tethers. A, whereas the free conformation of insulin is closed (left panel), upon receptor binding, detachment of the C-terminal B-chain β-strand is proposed to expose the hidden non-polar surface of the A-chain (right panel), including IleA2 and ValA3 (green). The aromatic side chain of PheB24 (red) may function as a structural switch. GlyA1 is shown as an open circle, and B29 and B30 are shown as purple and back circles, respectively. B, tethering the A- and B-chains was effected by bifunctional cross-linking (dashed blue line) between the ε-amino group of LysB29 (purple circle) and the α-amino group of GlyA1 (open circle). C, a histogram showing the relative (rel.) activity of tethered insulin analogs is plotted as a function of the number of atoms between the α-carbons of B29 and A1 (horizontal axis), including the B29 side chain and, when present, the side chain of d-LysA1 (magenta). Bar heights indicate receptor-binding affinities relative to wild-type human insulin. Black bars indicate linkage to the α-amino group of GlyA1, whereas magenta bars indicate linkage to the ε-amino group of d-LysA1. The asterisk indicates substantial relative activity (35 ± 3%) exhibited by NεA1,NεB29-ethylene glycol bissuccinoyl d-LysA1-insulin. D, shown is a schematic design of representative cross-linked insulin derivatives. Upper panel, NαA1,NεB29-ethylene glycol bissuccinoyl insulin; lower panel, NεA1,NεB29-ethylene glycol bissuccinoyl d-LysA1-insulin. In the upper construct, the total number of atoms between the α-carbons of B29 and A1 is the sum of the lengths of the linker (dashed blue line; 12 atoms) and the LysB29 side chain (magenta; 5 atoms) plus the Nα of GlyA1:12 + 5 + 1 = 18. In the lower construct, the tether is extended by the side chain of d-LysA1:12 + 5 + 5 = 22. E, shown is the design of three single-chain analogs. Upper panel, PIP, a 53-residue analog (SCI-53) with a dipeptide-connecting domain (AK); middle panel, SCI-57; lower panel, 58-residue analog (SCI-58) with connecting domain AAAAAAK. In SCI-57, B30 (Thr) and a 6-residue linker (sequence GGGPRR) interpose 23 atoms between the α-carbons of B29 and A1; the corresponding numbers of intervening atoms in PIP and SCI-58 are 11 and 26, respectively.
Although few SCIs have been characterized to date and no systematic studies have been reported (13, 16, 17, 30-33), decades-old literature describes the use of bifunctional chemical cross-linking reagents to tether the C terminus of the B-chain to the N terminus of the A-chain (Fig. 2B, blue dashed segment) (19, 34-37). The linkers were ordinarily attached to the ε-amino group of LysB29 and the α-amino group of GlyA1, thereby mimicking the effects of a connecting peptide. Because d-amino acid substitutions at A1 are well tolerated (38), an alternative design employed a d-LysA1 ε-amino group to extend the effective linker length (19). We may revisit the properties of these modified insulins as a foundation for the design of SCIs.
The activity of a series of B29-A1 cross-linked insulin analogs was evaluated by Nakagawa and Tager (19) in relation to the number of atoms (carbon or oxygen) in the linker (Fig. 2C). Whereas a 4-atom linker (NαA1,NεB29-succinoyl insulin) yielded an analog with an activity of 1.4 ± 0.5% relative to insulin and 5.2% relative to a bi-derivatized control, an analog containing a 12-atom linker (NαA1,NεB29-ethylene glycol bissuccinoyl insulin) exhibited an activity of 11 ± 3% relative to insulin and 40% relative to a bi-derivatized control. Including the LysB29 side chain, 18 atoms were interposed between the α-carbons of B29 and A1 (Fig. 2D) (19), analogous to a tether comprising ThrB30 and a 5-amino acid residue connecting domain (C-domain). Although reduced, the activity of NαA1,NεB29-ethylene glycol bissuccinoyl insulin is 100-fold higher compared with the porcine insulin precursor (PIP) (28), a 53-residue SCI containing a dipeptide linker (Fig. 1A) (14), and 1000-fold higher compared with an SCI in which LysB29 and GlyA1 are connected by a peptide bond (thereby interposing 2 atoms between respective α-carbons) (15, 17). Biophysical characterization of NαA1,NεB29-ethylene glycol bissuccinoyl insulin revealed an increase in its thermodynamic stability (ΔΔGu = 1.9 kcal/mol at 23 °C) and decreased flexibility, as indicated by an ∼103-fold retardation in the overall rate of amide proton exchange in D2O (39). Additional play between A- and B-chains was introduced via d-LysA1 to yield NεA1,NεB29-ethylene glycol bissuccinoyl d-LysA1-insulin. The activity of this construct was observed to be 35 ± 3% relative to insulin and 40% relative to a bi-derivatized control (Fig. 2C, asterisk) (19). Including the side chains of both LysB29 and d-LysA1, the number of atoms between the B29 and A1 α-carbons is 22 (Fig. 2D), analogous to interposition of ThrB30 and a 6-residue C-domain (SCI-57) (Fig. 2E). SCIs containing 7-residue C-domains have been described previously and exhibit relative receptor-binding activities of 14% (linker AAAAAAK) (18) and 28% (linker GGGPGKR) (32).
In this work, we describe the design, synthesis, and characterization of a novel monomeric SCI. A 6-residue glycine-rich linker is interposed between ThrB30 and GlyA1, which provides a spacing of 23 atoms between the B29 and A1 α-carbons (Fig. 2E, middle panel), thus predicted by the cross-linking studies of Tager and co-workers (19, 39) to confer partial but significant activity. The connecting segment (sequence GGGPRR) (Fig. 2E, blue) contains a central proline (boldface) to facilitate a chain reversal (31) and retains a dibasic element (underlined) at the CA junction. The insulin moiety contains four substitutions (one in the A-domain and three in the B-domain; ThrA8 → His, HisB10 → Asp, ProB28 → Asp, and LysB29 → Pro) collectively designed (i) to achieve electrostatic balance (40), (ii) to prevent self-assembly (41), (iii) to restore wild-type receptor-binding activity in compensation for the connecting segment (42, 43), and (iv) to optimize thermodynamic stability (44, 45). The corresponding two-chain insulin analog (designated 2CA) was prepared as a control to probe the effects of these substitutions in the absence of a connecting segment. The multistep design of the present 57-residue single-chain insulin analog (designated SCI-57) provides proof of principle for the rational engineering of an ultrastable insulin monomer.
SCI-57 was prepared by chemical synthesis using native fragment ligation (46), which is, to our knowledge, the first application of this methodology to a proinsulin analog. The synthetic protein exhibits a remarkable richness of structure and full receptor-binding activity. Surprisingly, two-dimensional NMR studies demonstrate non-local contacts between the artificial connecting segment and a native-like insulin moiety. The glycine-rich linker thus folds against the insulin moiety: its central pyrrolidine ring (designated ProC4; polypeptide position 34) contacts ValA3 at the edge of the hydrophobic core, whereas the final residue (ArgC6; polypeptide position 36) extends the A1-A8 α-helix (polypeptide residues 37-44).6 Comparison of SCI-57 and 2CA provides evidence that the connecting segment damps conformational fluctuations in the insulin moiety. As observed in studies of chemically cross-linked insulin derivatives (39), SCI-57 exhibits a marked increase in thermodynamic stability as probed by guanidine denaturation studies (ΔΔGu = 0.7 ± 0.1 kcal/mol relative to 2CA and 1.9 ± 0.1 kcal/mol relative to wild-type insulin). The striking functional and biophysical properties of SCI-57 validate the heuristic reasoning employed in its stepwise design. In addition, the feasibility of total synthesis promises to enable further studies of SCIs by nonstandard mutagenesis, including insertion of photoactivable amino acid derivatives (47) and inversion of chiral centers as stereospecific probes of conformational change (11, 21, 29, 48).
Protein engineering not only facilitates biophysical studies, but also enhances medical technologies. The coming decades face an emerging epidemic of diabetes mellitus, particularly in the developing world (49). The engineering of ultrastable proteins such as SCI-57 may thus have societal benefits in regions of Africa and Asia where electricity and refrigeration are not consistently available. Because degradation rates of insulin analogs correlate inversely with their relative stabilities (50, 51) and are further accelerated by conformational fluctuations (51), SCI-57 may represent a class of ultrastable analogs whose pharmaceutical formulation may enhance the safety and efficacy of insulin replacement therapy (52). We thus envisage that ultrastable single-chain formulations, designed in accord with the principles exemplified here, could advance the treatment of diabetes mellitus in the developing world (49, 53).
EXPERIMENTAL PROCEDURES
Materials—Human insulin was provided by Novo Nordisk (Copenhagen, Denmark). PIP was provided by Prof. Y.-M. Feng (Institute of Biochemistry, Academica Sinica, Shanghai, China). Sources of commercial reagents are provided under supplemental “Experimental Procedures.”
Synthesis of [HisA8,AspB10,AspB28,ProB29]Insulin—To obtain 2CA, chain combination (54) was effected by interaction of the S-sulfonated derivative of the A-chain analog (30 mg) and B-chain analog (15 mg) in 10 ml of 0.1 m glycine buffer (pH 10.6) in the presence of dithiothreitol (5.2 mg) as described (7, 55). Detailed synthetic methods are provided under supplemental “Experimental Procedures.” CM52-cellulose chromatography of each combination mixture enabled partial isolation of the hydrochloride form of the protein contaminated by free B-chain (combined weight of 4.3 mg). Final purification was accomplished by C18 reverse-phase high performance liquid chromatography (RP-HPLC), yielding 2.8 mg of the insulin analog. Re-chromatography by analytical RP-HPLC gave a single peak; its predicted molecular mass (5808.5 Da) was verified (5807.6 Da) by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. The overall yield of 2CA was similar to that obtained in control syntheses of wild-type human insulin.
Synthesis of Single-chain Insulin Analog—SCI-57 was synthesized by native chemical ligation (46) of three peptides: segment I (B1-B6; polypeptide residues 1-6), segment II (B7-A6; polypeptide residues 7-42), and segment III (A7-A21; polypeptide residues 43-57). Segments III and II were first ligated; their product was then ligated to segment I to yield the reduced and unfolded 57-residue polypeptide. This synthetic scheme, exploiting cysteines at B7 and A7 as ligation sites, thus employs peptides of nominal lengths 6, 36, and 15 residues exclusive of C-terminal thioester extensions (segments I and II; see below). Assembly of peptide segments I and II employed t-butoxycarbonyl chemistry and an in situ neutralization protocol as described (56); peptide segment III was assembled by Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry on polyethylene glycol-Polystyrene graft polymer (PEG-PS™). Each segment was synthesized by a manual solid-phase protocol as described under supplemental “Experimental Procedures.” Analyses of purity by RP-HPLC and subsequent verification of peptide masses were performed using an Agilent 1100 liquid chromatography/mass spectrometry system. Preparative HPLC was performed using C4 or C8 columns (1.0 × 25 or 2.2 × 25 cm).
Redox-coupled Protein Folding—Folding of the reduced SCI-57 polypeptide was initiated by addition of a redox-coupled buffer (0.4 ml) consisting of 100 mm GSH and 10 mm GSSG; the pH was adjusted to 8.6 with 2 n NaOH. This solution was immediately diluted with distilled deionized water to 40 ml. The extent of folding was periodically monitored by liquid chromatography/mass spectrometry. When almost no further folding was observed, the mixture was acidified to pH 2 with 10% trifluoroacetic acid, and acetonitrile was added to a final concentration of 10% (v/v). Three successive folding reactions were undertaken as described under supplemental “Experimental Procedures”; the solutions were subjected to RP-HPLC using a C4 column. In total, 6.6 mg was obtained, representing an overall yield of 24.4% for the two-step ligation procedure, subsequent folding, and purification. Mass spectrometry of the final product gave an observed molecular mass (6370.8 Da), in accord with the predicted molecular mass (6371.2 Da).7
Receptor Binding Assays—Relative activity is defined as the ratio of analog to wild-type human insulin required to displace 50% of specifically bound human [125I-TyrA14]insulin. A human placental membrane preparation containing the IR was employed as described (29). In all assays, the percentage of tracer bound in the absence of competing ligand was <15% to avoid ligand depletion artifacts. Assays were repeated in duplicate three times.
Circular Dichroism—Far-UV CD spectra of each analog were obtained using an Aviv spectropolarimeter equipped with a thermister temperature control and an automated titration unit for guanidine denaturation studies. CD samples for wavelength spectra contained 25-50 μm insulin analog in 50 mm KCl and 10 mm potassium phosphate (pH 7.4); samples were diluted in the same buffer to 5 μm for equilibrium denaturation studies. Data were obtained at 37 °C. Guanidine denaturation data were fitted to a two-state model by nonlinear least square regression (57).
NMR Spectroscopy—Spectra were obtained at 700 MHz with a cryogenic 1H NMR probe as described (7); the protein concentration was 0.5 mm. Resonance assignment was based on two-dimensional nuclear Overhauser enhancement (NOE) spectroscopy (NOESY; mixing times of 150 and 80 ms for SCI and 180 and 80 ms for 2CA), total correlation spectroscopy (TOCSY; mixing times of 30 and 55 ms), and double-quantum filtered correlated spectroscopy spectra. Three solution conditions were used: 1) in aqueous solution at pH 7.4 and 25 °C, 2) at pH 8.0 and 32 °C, and 3) in 20% (v/v) deuterioacetic acid at pH 1.9 and 25 °C. The acetic acid co-solvent facilitates analysis of exchangeable amide resonances, otherwise incomplete at neutral pH because of base-catalyzed solvent exchange and conformational broadening (7); in this co-solvent, insulin retains a native-like monomeric fold. Helix-related and interdomain hydrogen bonds were inferred from the pattern of protected amide resonances in D2O solution containing 20% deuterioacetic acid (58).
Structure Calculations—Distance geometry (DG)/simulated annealing calculations were performed using the program DG-II (17); restrained molecular dynamics calculations were performed using X-PLOR (59). NOE-related and dihedral angle restraints were used for molecular modeling. NOEs were classified as strong (<2.5 Å), medium (<3.3 Å), or weak (<5.0 Å) in reference to standard intra-residue cross-peaks. Distance-bound corrections were made for methyl groups and methylene protons for which stereospecific assignments could not be obtained. A reference structure of an engineered insulin monomer is provided by DKP-insulin (two-chain insulin analog containing three substitutions in the B-chain (AspB10, LysB28, and ProB29); Protein Data Bank code 2jmn) (7, 23).
Proteomics Tools—Molecular masses and pI values were obtained based on protein sequence using the Compute pI/Mw suite of software tools on the ExPASy Proteomics Server (ca.expasy.org/tools) with the pI algorithm (ca.expasy.org/tools/protparam.html).
RESULTS
Our results are presented in three parts. We first provide an overview of protein design, emphasizing both its theoretical foundation (60) and links to classical studies of insulin analogs (41-45, 61, 62). We next describe the synthesis and biochemical characterization of SCI-57, verifying design goals regarding activity and stability. Finally, the solution structure of SCI-57 is presented. As expected, SCI-57 retains a native-like insulin fold. Surprisingly, however, the connecting segment exhibits a non-random conformation and exerts a transmitted effect on the amplitude of inter-residue NOEs in the insulin moiety. Such effects provide a model for the non-local damping of conformational fluctuations in a globular protein (63).
Design of a Novel Single-chain Analog
SCI-57 incorporates four intended design elements, described in turn below. Although in each case a rationale is provided, such arguments must be considered as heuristic given the limited availability of SCI structures (15, 33, 64).8 Verification of predicted features is described in the subsequent sections.
Connecting Peptide Sequence and Length—The foreshortened connecting segment contains a glycine-rich linker with a central proline (GGGPRR; positions C1-C6), predicted to favor a flexible chain reversal. A dibasic element is retained at the CA junction to mimic the pattern of charges in proinsulin. The length of this segment is similar to that of a d-LysA1-extended B29 chemical cross-link, which was observed to be compatible with partial activity (11 ± 2% relative to wild-type insulin) and which conferred enhanced stability (ΔΔGu = 1.9 kcal/mol) (19, 39).
Compensation between Favorable and Unfavorable Receptor-binding Elements—Because the 6-residue linker was expected to impair activity by at least 3-fold, two substitutions were introduced in the insulin moiety (ThrA8 → His and HisB10 → Asp), previously shown to enhance the activity of two-chain analogs by at least 3-fold (42). These substitutions also enhance the thermodynamic stability of insulin (44, 45). Such enhancement has a theoretical foundation. Based on rules derived from studies of model peptides (65), HisA8 would increase the C-terminal end propensity (C-Cap) of the A1-A8 α-helix by replacing an unfavorable β-branched amino acid. Similarly, the negative charge of AspB10 would be expected to interact more favorably than HisB10 with the electrostatic dipole of the B-chain α-helix (66, 67), an effect that could be augmented by side chain/main chain hydrogen bonding (68).
Prevention of Dimerization and Higher Order Assembly—Because the complex self-association properties of insulin and proinsulin ordinarily confound biophysical studies, a monomeric SCI was sought. Whereas the AspB10 substitution in insulin blocks zinc binding and an associated trimer-forming surface (41), substitutions at B28 and B29 impair dimerization (41, 43). We thus employed the substitutions ProB28 → Asp (as in Novalog®, a rapid-acting insulin analog in clinical use) (33) and LysB29 → Pro (as in Humalog®, another analog formulation) (69).
Electrostatic Balance—Because the solubility of insulin and insulin analogs depends critically on pH in the range 5-7 due to isoelectric precipitation (51), it seemed desirable to maintain a pI similar to or lower than that of wild-type insulin (predicted pI 5.4), thereby preserving or enhancing solubility of the single-chain analog at neutral pH. The linker sequence and substitutions in the insulin moiety were thus chosen to maintain a balance between net changes in positive and negative charges. The positive charges of the dibasic CA junction and potential positive charge of HisA8 are offset by AspB10, AspB28, and the removal of a positive charge at B29. The connecting segment is associated with the near-balanced removal at neutral pH of the C-terminal carboxylate of the B-chain and N-terminal amino group of the A-chain. The calculated pI of SCI-57 (5.0) is thus 0.4 units lower than that of zinc-free insulin.
Synthesis and Characterization
The total chemical synthesis of SCI-57 was accomplished by the native ligation of three peptide segments (see “Experimental Procedures”). This protocol circumvents the cumulative inefficiency of sequential peptide synthesis (46). An overall yield of almost 25% (including losses at each ligation step, purification of intermediates, and redox-coupled folding) was achieved. In addition to the advantages of native fragment ligation in preparing the unfolded polypeptide, correct redox-coupled disulfide pairing by SCIs (30) is generally more efficient than that obtained by insulin chain combination (54, 55).9 Realizing this potential efficiency in SCI-57 required iterative cycles of redox-coupled folding due to the unfavorable solubility properties of the reduced and unfolded polypeptide (see supplemental “Experimental Procedures”). The molar incorporation of the A-chain in the synthesis of [HisA8,AspB10, AspB28,ProB29]insulin was only 8%, a value similar to (but slightly less than) that observed in the synthesis of wild-type insulin (70).10
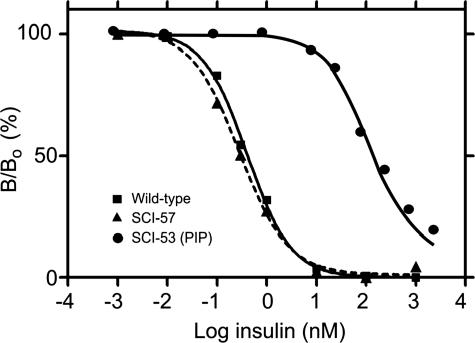
The receptor-binding activity of SCI-57, measured using a human placental membrane preparation, is 130 ± 8% relative to that of wild-type human insulin (Fig. 3, ▴ and ▪, respectively). Under assay conditions, the base-line affinity of wild-type insulin for the IR was observed to be 0.39 ± 0.03 nm. Control studies of PIP indicated a relative affinity of 0.23 ± 0.04% (Fig. 3, •), in accord with past studies. The affinity of 2CA is >400% (data not shown); accurate assessment of its enhanced activity would require use of a radiolabeled superactive analog as tracer. Partial hindrance to receptor binding by the 6-residue C-domain in SCI-57 is thus offset by the favorable substitutions in its modified insulin moiety. Such functional balance between design elements is in accord with compensation between favorable and unfavorable single amino acid substitutions in conventional two-chain insulin analogs (27, 42). Although the AspB10 substitution has been associated with increased mitogenicity and cross-binding to the IGF receptor, such effects are not significant in SCI-57, presumably due to its C-domain.11
FIGURE 3.
IR binding assay. Representative displacement data illustrate the activity of the present single-chain analog with a 6-residue linker (SCI-57; ▴), human insulin (▪), and PIP (an SCI-53; •). Whereas binding of SCI-53 (PIP) is reduced by >500-fold, a slight enhancement of affinity by SCI-57 is seen as a leftward shift of its displacement curve. The y axis (B/Bo) indicates percent receptor-bound 125I-labeled human insulin; the x axis (logarithmic scale to base 10) indicates the concentration of unlabeled competing insulin analog.
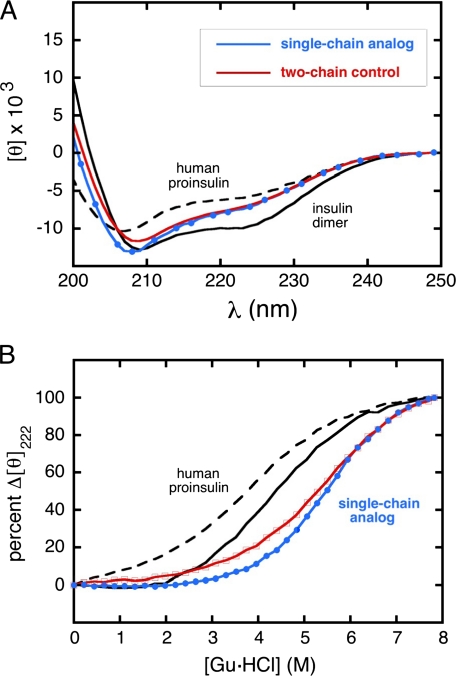
Initial insight into the structure of SCI-57 was provided by far-UV CD spectroscopy (Fig. 4A). Reference spectra are provided by wild-type insulin (solid black line) and proinsulin (dashed black line) under conditions in which these proteins are predominantly dimeric (71). Because the CD spectra are normalized by mean residue ellipticity, the spectrum of proinsulin appears less helical than that of insulin (attenuated minima at 208 and 222 nm and attenuated maximum below 200 nm); this is due to the random coil and turn contributions of the 35-residue C-domain. SCI-57 and 2CA exhibit native-like CD spectra with slight attenuation of helix-related features, presumably due to the absence of self-assembly (27). Relative to 2CA, the presence of the C-domain is not associated with a CD-detectable perturbation. The similarity between the CD spectra of SCI-57 and 2CA stands in contrast to the marked (and poorly understood) differences between the spectra of wild-type insulin and the 53-residue single-chain analog PIP (18).
FIGURE 4.
Far-UV CD spectra of insulin, proinsulin, and analogs. A, spectra of single- and two-chain insulin analogs (SCI-57 and 2CA): human insulin (solid black line), proinsulin (dashed black line), SCI-57 (blue line), and 2CA (red line)). Ellipticity was observed at each nanometer step; in the spectrum of the single-chain analog, 40% of data points are shown (blue circles). Whereas the analogs are monomeric, human insulin is predominantly dimeric (in equilibrium with hexamer) under these conditions; such assembly enhances CD-detected helical features. Spectra are normalized to provide mean residue ellipticity ([θ]); whereas insulin and proinsulin exhibit similar α-helix content, in such a normalized spectrum, the helix-related amplitude at 222 nm of proinsulin is attenuated due to the random-coil contribution of the connecting peptide. An analogous effect of normalization yields similar values of mean residue ellipticity at 222 nm for SCI-57 and 2CA, whereas the helix-related ellipticity of the SCI per molecule is enhanced by ∼10%. B, CD-detected guanidine denaturation studies as monitored at 222 nm. The color code is as described for A. All data points in titration of analogs are shown (red squares and blue circles). The extreme rightward shift of the denaturation transition of the single-chain analog relative to human proinsulin reflects its markedly augmented thermodynamic stability. Data in each panel were obtained at 37 °C in 10 mm potassium phosphate buffer (pH 7.4). Gu·HCl, guanidine hydrochloride.
The thermodynamic stability of SCI-57 was inferred from CD-detected protein denaturation (Fig. 4B). Studies were conducted at 37 °C to simulate the storage of unrefrigerated insulin formulations in the developing world. A marked rightward shift was observed in the unfolding transition of SCI-57 as a function of guanidine concentration (Fig. 4B, blue line) relative to 2CA (red line), wild-type insulin (solid black line), and proinsulin (dashed black line). Thermodynamic parameters extracted from a two-state model (57) are given in Table 1. Whereas the inferred free energy of unfolding (ΔGu) of human insulin at neutral pH and 37 °C is 2.4 ± 0.1 kcal/mol, the stability of 2CA is higher (ΔGu = 3.6 ± 0.1 kcal/mol), presumably due to the stabilizing effects of the HisA8 and AspB10 substitutions (44, 45). A further increase in stability is provided by the connecting segment (ΔGu = 4.3 ± 0.05 kcal/mol). This gain in stability occurs despite similarity of their normalized CD spectra (Fig. 4A).
TABLE 1.
Thermodynamic stabilities of insulin analogs
Stabilities were inferred from CD-detected guanidine denaturation studies at 37°C as described (57).
| Analog | ΔGu | Cmid | m |
|---|---|---|---|
| kcal/mol | m | kcal/mol/m | |
| Human insulin | 2.4 ± 0.1 | 4.1 ± 0.2 | 0.57 ± 0.02 |
| AspB28-insulin | 2.4 ± 0.1 | 4.1 ± 0.1 | 0.60 ± 0.01 |
| 2CAa | 3.6 ± 0.1 | 5.8 ± 0.2 | 0.62 ± 0.02 |
| SCI-57b | 4.3 ± 0.1 | 5.5 ± 0.1 | 0.79 ± 0.02 |
| Proinsulin | 2.8 ± 0.1 | 4.0 ± 0.2 | 0.71 ± 0.03 |
2CA (two-chain analog of 51 residues) contains substitutions ThrA8 → His, HisB10 → Asp, ProB28 → Asp, and LysB29 → Pro.
SCI-57 is single-chain insulin analog with a 6-residue C-domain (GGGPRR) and the same four substitutions as in 2CA.
Our observation of a linker-associated enhancement of thermodynamic stability stands in contrast to the absence of such enhancement in comparison of wild-type insulin and PIP (ΔΔGu = 0 ± 0.2 kcal/mol) (72). The increased stability of SCI-57 at 37 °C (ΔΔGu = 0.7 ± 0.2 kcal/mol relative to 2CA and ΔΔGu = 1.9 ± 0.2 kcal/mol relative to wild-type insulin) is nonetheless consistent with the increased stability of the B29-A1 cross-linked insulin derivative at 23 °C (NαA1,NεB29-bissuccinoyl insulin; ΔΔGu = 1.9 kcal/mol relative to wild-type insulin), as was also measured by guanidine denaturation (39). The variable extent of augmented stability among SCIs or cross-linked derivatives of different tether lengths (between -0.2 and +2.0 kcal/mol, including the uncertainty) presumably depends on an interplay between unfavorable strain (as might be introduced by the 2-residue mini-C-domain in PIP) and favorable effects of an intermediate-length linker on structure, dynamics, and solvation.
Solution Structure and Dynamics
1H NMR spectra of SCI-57 and 2CA exhibit at neutral pH and 25 °C a pattern of chemical shift dispersion and line widths similar to those observed in previous studies of engineered two-chain insulin monomers (7, 9). Whereas 1H NMR spectra of wild-type insulin and proinsulin under these conditions exhibit a broad envelope of poorly resolved resonances (due to both self-association and conformational exchange) (73-75), spectra of engineered insulin monomers typically exhibit motional narrowing and lack of dependence on protein concentration (in the range of 0.1-1 mm) characteristic of a 6-kDa monomer (7, 9). The favorable NMR features of SCI-57 in aqueous solution at neutral pH indicate that its intended monomeric framework (due to AspB10 and the AspB28-ProB29 element) was realized, thus generalizing the properties of two-chain analogs to the present single-chain context.
One-dimensional 1H NMR spectra of SCI-57 and 2CA are shown in Fig. 5 (A and C). Spectra were also obtained in 20% deuterioacetic acid (pH 1.9) (Fig. 5, B and D) to facilitate analysis of exchangeable 1H NMR resonances (58). The partial aliphatic character of deuterioacetic acid weakens the hydrophobic effect, preventing dimerization by the exposed non-polar surface of insulin; in this co-solvent, insulin retains a native-like structure (6) but exhibits reduced overall chemical shift dispersion relative to an engineered monomer at neutral pH (7), presumably due to more flexible packing within the hydrophobic core. SCI-57 retains a high quality spectrum in 20% deuterioacetic (Fig. 5B)12 despite the likely protonation (and hence neutralization) of AspB10 and AspB28 at pH 1.9. Negligible self-assembly of SCI-57 under either NMR condition was confirmed by the absence of characteristic dimer- or trimer-specific NOEs at potential interfaces (see below) (8, 10).
FIGURE 5.
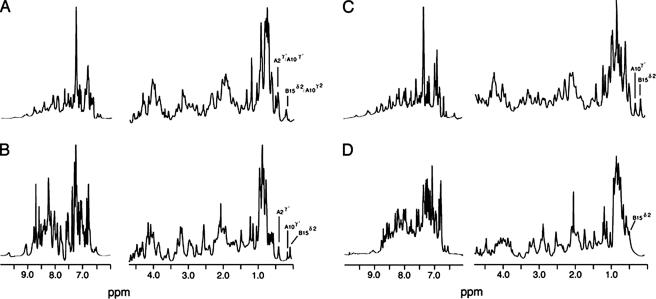
1H NMR spectra of corresponding single- and two-chain insulin analogs. A and B, spectra of SCI in aqueous solution (pH 7) and in 20% (v/v) deuterioacetic acid (pH 1.9), respectively; C and D, corresponding spectra of 2CA in aqueous solution (pH 7) and in 20% deuterioacetic acid, respectively. Spectra were obtained in each case at 25 °C at a protein concentration of ∼0.5 mm. Assignments of selected upfield-shifted aliphatic resonances are as labeled (0-1 ppm). In SCI, the large secondary shift of LeuB15 δ2-CH3 at neutral pH is maintained in 20% deuterioacetic acid, whereas in 2CA, this and other upfield methyl shifts are attenuated by the organic co-solvent. The overall envelope of exchangeable resonances at pH 7 (7-10 ppm) is attenuated relative to acidic pH due to base-catalyzed solvent exchange and conformational broadening. Spectra were obtained at 700 MHz.
Complete sequential resonance assignment of SCI-57 and 2CA was obtained by standard homonuclear methods (see supplemental “Experimental Procedures,” Figs. S4 and S5, and Table S3 and S4). Similar patterns of chemical shifts are seen in the two proteins. As expected, the largest changes in chemical shift are observed adjoining the connecting segment (B27-B30 and A1-A3). Small differences that are widely distributed in the structure of the insulin moiety are nonetheless observed (see supplemental “Experimental Procedures” and Table S5). Such differences are illustrated by the upfield-shifted aliphatic resonances between 0 and 1 ppm. Whereas in each case the most upfield methyl resonance is assigned to LeuB15, the precise position of this resonance and other members of the B15 spin system varies (Fig. 5, A and C). Such differences may reflect small changes in the orientation of neighboring aromatic rings (TyrA19, PheB24, and TyrB26). Previous ring current shift simulations have highlighted the marked sensitivity of such aliphatic chemical shifts to ring orientation (8, 76). NOESY spectra of SCI-57 and 2CA nonetheless exhibit similar patterns of contacts between aromatic and aliphatic spin systems (Fig. 6). Whereas (as above) 1H NMR spectra of insulin and monomeric insulin analogs in 20% deuterioacetic acid typically exhibit reduced dispersion, including in the upfield region (Fig. 5D) (7), the spectrum of SCI-57 retains secondary shifts similar in magnitude to those observed at neutral pH (exemplified by the methyl resonances of IleA2, IleA10, and LeuB15 in Fig. 5B). These features suggest that the connecting segment enables SCI-57 to resist in part the destabilizing effects of the co-solvent on non-polar packing interactions.
FIGURE 6.
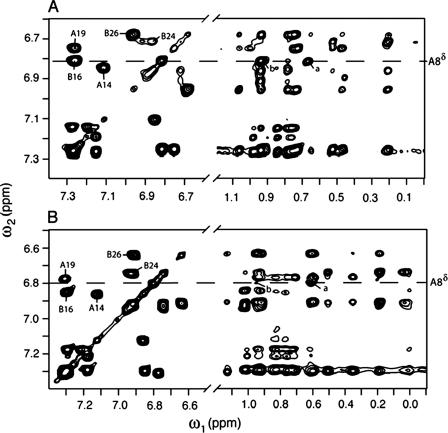
Two-dimensional NMR spectra of corresponding single- and two-chain insulin analogs. A, TOCSY spectrum of SCI at pD 7.6 and 32 °C (direct meter reading) containing aromatic cross-peaks (left panel) and corresponding NOESY spectrum containing long-range NOEs between aromatic and aliphatic protons (right panel). Contacts between HisA8 Hδ and the γ-CH3 of ValA3 are labeled a and b, respectively. B, corresponding spectral regions of the parent two-chain analog. The two proteins exhibit similar overall trends with only subtle differences. Analogous A8-A3 contacts are labeled a and b. Spectra were obtained at 700 MHz.
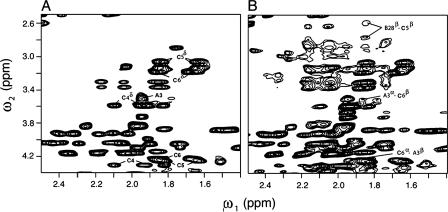
The 1H NMR spectrum of SCI-57 contains six additional spin systems (relative to 2CA) due to the C-domain as illustrated in Fig. 7A. For clarity, respective linker residues GGGPRR are designated C1-C6 (residues 31-36 in the intact polypeptide). Despite its central proline, no evidence is observed for cis,trans-GlyC3-ProC4 isomerization. The Hδ resonance of ProC4 exhibits NOEs to each methyl group of ValA3 (Table 2), providing evidence for the non-polar packing of the pyrrolidine ring. That the engineered C-domain exhibits a non-random conformation is suggested by observation of multiple additional long-range NOEs involving C4-C6 (PRR) (Table 2). These indicate contacts to side chains in or adjoining the hydrophobic core of the insulin moiety (IleA2, ValA3, TyrA19, and TyrB26); representative contacts are illustrated in Fig. 7B. Structural organization within the C-domain is indicated by a medium-strength (i, i+3) NOE from GlyC1 to ProC4, reflecting a chain reversal. These and other inter-residue NOEs in SCI-57 and 2CA are summarized by diagonal plot in Fig. 8.
FIGURE 7.
Linker-specific two-dimensional NMR signals in single-chain insulin analog. A, region of the TOCSY spectrum of SCI containing α-β and β-δ or γ-δ peaks of residues PRR in GGGPRR (C1-C6; polypeptide residues 31-36); B, related inter-residue contacts in the NOESY spectrum provide evidence for long-range packing by the dibasic element: ArgC5-AspB28 and ArgC6-ValA3. Spectra were obtained at 700 MHz.
TABLE 2.
Linker-related NOEs in 1H NMR spectra of SCI-57
The B-, C-, and A-domains of SCI-57 comprise respective segments 1-30, 31-36, and 37-57 of the single-chain polypeptide. In addition to NOEs from the connecting domain to the A- and B-domains, an (i, i+3) turn-related NOE is observed within the linker (GlyC1 HN-ProC4 Hα).
| To A-domain | To B-domain |
|---|---|
| ProC4 Hδ-ValA3 γ1,2-CH3 | GlyC3 Hα-TyrB26 Hδ |
| ArgC5 Hα-IleA2 HN | ArgC5 Hα-AspB28 Hβ |
| ArgC5 Hα-ValA3 Hβ | ArgC5 Hβ-AspB28 Hβ |
| ArgC5 Hδ-IleA2 δ-CH3 | ArgC5 Hγ-TyrB26 Hδ |
| ArgC5 Hγ-TyrA19 Hδ,ε | |
| ArgC6 Hα-ValA3 Hβ | |
| ArgC6 Hα-ValA3 γ1,2-CH3 | |
| ArgC6 Hβ-ValA3 Hα |
FIGURE 8.
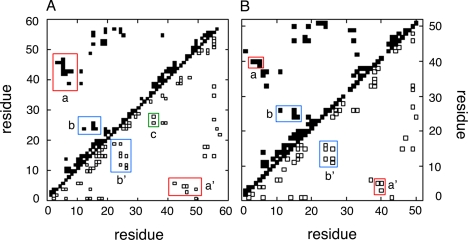
Diagonal plots of inter-residue NOEs in single- and two-chain insulin analogs. A, SCI-57; B, 2CA. More inter-residue NOEs are observed within the insulin moiety of the SCI-57 than in 2CA. Contacts between main chain protons are shown at lower right; side chain-side chain and side chain-main chain contacts are shown at upper left. Red boxes contain contacts from the B3-B5 segment to side chains of A9 and A10 (labeled as a or a′); contacts in blue boxes (b and b′) represent NOEs from the B24-B26 segment to the central B-chain α-helix residue diagnostic of native-like B-chain super-secondary structure. The green box (c in A) contains linker-related NOEs. In SCI, residues 1-57 indicate (i) B1-B30 (residues 1-30), (ii) linker (C1-C6; peptide positions 31-36), and (iii) A1-A21 (residues 37-57). In 2CA, residues 1-51 designate the B-chain (residues 1-30) and A-chain (residues 31-51).
The dibasic element at C5 and C6 exhibits (i, i+3) and (i, i+4) NOEs (Table 2), extending the pattern of α-helix-related NOEs in the classical A1-A8 segment. ArgC5 exhibits contacts to IleA2 and ValA3 (C5 Hα-A2 HN, C5 Hα-A3 Hβ, and C5 Hδ-A2 δ-CH3), and ArgC6 likewise contacts ValA3 (C6 Hα-A3 Hβ, C6 Hα-A3 γ1,2-CH3, and C6 Hβ-A3 Hα). This extended helical pattern leads to subtle changes in the conformation of HisA8, one of the stabilizing substitutions employed in the design of SCI-57. In both SCI-57 and 2CA, HisA8 exhibits expected (i, i+3) and (i, i+4) α-helix-related NOEs, but with differences in their details. Whereas both proteins share canonical dαN(i, i+3) A4-A8 and dαN(i, i+3) A5-A8 contacts, SCI-57 exhibits an A4 Hα-A8 Hα NOE, and 2CA exhibits an A5 Hα-A8-Hα NOE. SCI-57 (but not 2CA) exhibits NOEs from A8 Hβ to the side chains of GluA4 (Hγ) and GlnA5 (Hβ). The orientation of the A8 imidazole ring likewise differs as indicated by distinct patterns of NOEs to ValA3 and GluA4 from respective Hδ and Hε protons in the heterocycle (see supplemental “Experimental Procedures” and Table S1). Although the functional significance of these details is unclear, they illustrate how a foreshortened C-domain can lead to a transmitted change in a local side chain conformation.
Inspection of the diagonal plots in Fig. 8 reveals that SCI-57 exhibits significantly more inter-residue NOEs within the insulin moiety than were observed in the NOESY spectrum of 2CA. These include additional α-helix-related (i, i+3) and (i, i+4) contacts within each of the three canonical helical segments (comprising B9-B19, A1-A8, and A12-A18), long-range NOEs within the B-domain characteristic of its super-secondary structure (LeuB6 Hα-AlaB14 Hβ and CysB19 Hβ-GlyB23 HN), long-range NOEs within the A-domain (CysA6 HN-IleA10 γ′-CH3 and CysA6 Hα-IleA10 HN), and contacts between the B- and A-domains. The latter include multiple contacts involving the three cysteines (CysB19 Hα-GluA17 Hα, CysB19 Hα-AsnA21 HN, CysA6 Hα-HisB5 Hβ, CysA7 Hα-HisB5 Hβ, CysA7 Hβ-GlyB8 HN, and CysA11 Hα-HisB5 Hα) or neighboring residues (GlnB4 NεH-LeuA13 δ1,2-CH3, ValB18 γ1-CH3-LeuA16 δ1,2-CH3, and PheB25 HN-TyrA19 Hβ). In toto, the spectrum of SCI-57 exhibits 36 inter-residue NOEs not observed in the spectrum of 2CA; by contrast, the spectrum of 2CA contains only two NOEs (ValA3 γ2-CH3-TyrA19 Hε and AsnA21 Hδ-GlyB23 Hα) that are not observed in SCI-57. In each case, the differential NOEs are consistent with crystallographic protomers of wild-type insulin, and their presence or absence cannot be accounted for by technical differences in resonance line width or overlap. Because the NOESY spectra of SCI-57 and 2CA exhibit similar signal-to-noise ratios for intra-residue NOEs constrained by covalent structure (such as the ortho,meta aromatic NOEs of Phe and Tyr), observation of SCI-specific inter-residue NOEs reflects their attenuation in the spectrum of 2CA.
The difference in NOE density between SCI-57 and 2CA reflects an unusual structural richness in SCI-57; the 1H NMR spectrum of 2CA is consistent with spectra of other two-chain monomeric analogs (7, 9). Indeed, the anomalous attenuation of NOEs in the spectrum of 2CA relative to SCI-57 is reminiscent of previously described discrepancies between the spectra of insulin and insulin analogs relative to simulated NOESY spectra predicted by crystallographic protomers, attributed to conformational fluctuations (8, 77). Restoration of such “missing” NOEs has been observed upon engagement of wild-type insulin within the rigid R6 zincphenol hexamer (8). Whereas the spectrum of the hexamer also contains intersubunit NOEs across the dimer and trimer interfaces (8, 10), SCI-57 exhibits a richness of inter-residue NOEs without evidence of intermolecular contacts. These results suggest that the connecting segment constrains conformational fluctuations within and between the A- and B-domains in a manner that in part resembles damping of such fluctuations by protein assembly.
The structure of SCI-57 was calculated by DG/restrained molecular dynamics based on 852 restraints (783 NOEs, 47 dihedral angle restraints, and 22 hydrogen bond-related restraints) (see supplemental Table S6). The number of restraints per residue is thus 14.9 (SCI-57). Statistical information is provided in supplemental Table S6. The structure is essentially identical to native human insulin, in accord with their native-like patterns of inter-residue NOEs. Within the central well ordered region of the structure, root mean square deviations relative to the mean structure are in each case <0.3 Å (main chain atoms in B4-B24 and A2-A19) and <0.6 Å (side chain atoms in the same regions).
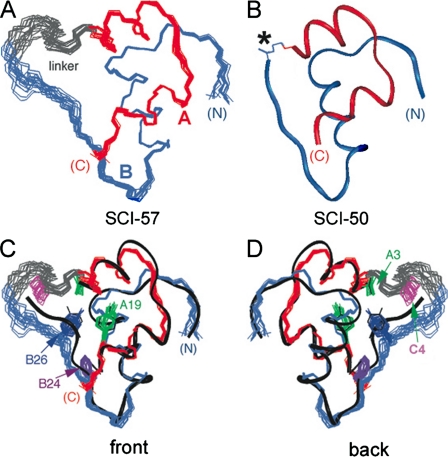
In the ensemble of DG/restrained molecular dynamics models of SCI-57 (Fig. 9A), the A- and B-domains (red and blue, respectively) resemble a crystallographic protomer of des-B30-[B29-A1]SCI-50 (Fig. 9B) (15). Alignment of the SCI-57 ensemble with a ribbon model of DKP-insulin (black) (7) is shown in Fig. 9, C (front view) and D (back view); selected side chains are shown (ValA3 (green), TyrA19 (green), PheB24 (purple), TyrB26 (dark blue), and linker residue ProC4 (magenta)). The foreshortened C-domain (shown in gray in Fig. 9) is partially ordered, especially at the PRR element (C4-C6), in accord with NOE patterns. This element extends the A1-A8 α-helix and stably folds against a hydrophobic crevice between the A- and B-domains, sealing one edge of the hydrophobic core. An α-helical hydrogen bond (not imposed as a restraint) is present between the carbonyl oxygen of ArgC6 and the peptide NH of ValA3. ProB29, ThrB30, and the GGG segment (C1-C3) are less well ordered but contribute to the shielding of hydrophobic surfaces in the insulin moiety that would otherwise be partially exposed to solvent.
FIGURE 9.
Solution structure of SCI. A, ensemble of DG/restrained molecular dynamics models of SCI (Protein Data Bank code 2jzq). The A-chain is shown in red, the B-chain in blue, and the linker in gray. Structures were aligned according to the main chain atoms of A2-A7, A13-A19, and B9-B26. B, ribbon model of des-ThrB30-SCI-50, an inactive 50-residue single-chain analog in which a peptide bond links LysB29 to GlyA1 (Protein Data Bank code 1pid) (15). C and D, front and back views, respectively, of SCI-57 with selected side chains shown relative to the ribbon model of DKP-insulin (black) (Protein Data Bank code 2jmn) (7). The side chain coloring scheme is as follows: ValA3, green; TyrA19, green; PheB24, dark purple; TyrB26, blue; and linker residue ProC4, magenta.
DISCUSSION
Structure-function relationships in insulin have been inferred from patterns of sequence conservation (4) and extensively probed by mutagenesis (4, 20, 21, 24, 25, 55, 78-81). This study was motivated by anomalies encountered in past studies of single-chain insulin analogs (5, 17). Although native-like in conformation, the presence of mini-C-domains markedly impedes receptor binding. These and other observations support a model in which the C-terminal β-strand of the B-chain (B24-B28) detaches from the hydrophobic core of insulin to insert between domains of the IR (82).
Experimental design builds on classical studies of chemically cross-linked insulin derivatives (19, 34-37) to obtain a single-chain analog containing a 6-residue C-domain (SCI-57). Because this linker would be expected to permit partial receptor binding (with affinity reduced by 3-10-fold relative to wild-type insulin) (18, 32), substitutions shown previously to enhance receptor binding (HisA8 and AspB10) (42, 45, 61, 83) were introduced into the insulin moiety. Additional substitutions were employed at B28 and B29 to prevent dimerization and hence enable rigorous spectroscopic studies of the analog as a monomer in solution. The B28-B29 element (AspB28-ProB29) resembles that employed in the clinical rapid-acting analog Humalog® (insulin lispro; LysB28-ProB29) (33, 43) except that the positive charge of LysB28 is replaced by the negative charge of AspB28, a substitution employed in the clinical analog Novalog® (insulin aspart; AspB28-LysB29) (41). This charge substitution was introduced to maintain electrostatic balance with respect to the dibasic element in the foreshortened C-domain (ArgC5-ArgC6; polypeptide positions 35 and 36) and HisA8 substitution. The net result of such protein engineering is to obtain a monomeric SCI that is highly soluble at neutral pH and retains native receptor-binding activity (130 ± 8% relative to human insulin).
SCI-57 Synthesis by Native Chemical Ligation—Pharmaceutical manufacture of insulin is accomplished by recombinant expression of precursor polypeptides in microorganisms (84). By contrast, in this study, we employed native fragment ligation (46) to enable rapid and efficient synthesis of SCI-57. This methodology circumvents the successive inefficiency of standard solid-phase synthesis, which ordinarily limits the feasibility of polypeptides >45 residues with sequences rich in cysteine and non-polar side chains. Such limitations were encountered in a previous sequential synthesis of a 50-residue SCI (17). The power of native fragment ligation to overcome these limitations has been described recently in studies of insulin-like growth factor (IGF) I (85). The overall yield of SCI-57 is limited by aberrant aggregation and precipitation of intermediates encountered during redox-coupled folding of the reduced polypeptide (see supplemental “Experimental Procedures”). Analogous inefficiencies have been described in the in vitro refolding of proinsulin at high polypeptide concentration (86).
The development of total synthetic access (although not required for the preparation of conventional analogs) promises to enable the incorporation of nonstandard amino acids. The utility of nonstandard mutagenesis in structure-function studies of insulin has been demonstrated by inversion of chiral centers (11, 21, 23, 25, 26) and the introduction of photoactivable side chain derivatives to map hormone-receptor contacts (23, 47). A key advantage of a synthetic SCI framework for photocross-linking studies would be the feasibility of simultaneously introducing photoprobes in the A- and B-domains to test the hypothesis that an individual insulin molecule binds in trans to the two α-subunits of the insulin holoreceptor (87, 88). Extension of site-specific photocross-linking studies to the homologous single-chain ligands IGF-I and IGF-II (85) promises to illuminate the similarities and differences between the modes of binding of insulin and IGFs to cognate receptor tyrosine kinases (89). Efforts to extend the present synthetic protocol to enable such applications are in progress.
Activity of Single-chain Analogs—SCI-57 provides an example of a single-chain insulin analog with native activity. Human proinsulin itself, which contains a 35-residue C-domain, binds only weakly to the IR. The structural basis of such low affinity (1-2% relative to insulin) is not well understood, as proinsulin contains a native-like insulin moiety with a disordered connecting segment (71, 74). Furthermore, the connecting peptide does not obstruct native self-assembly (90). Because the self-association surfaces of insulin overlap its receptor-binding surface (12, 78), these observations suggest that proinsulin likewise retains a functional receptor-binding surface. Given that insulin binds within an internal crevice of an inverted V-shaped ectodomain (91), it is possible that the large C-domain cannot be accommodated without an unfavorable distortion.13
The molecular basis of the low activity of proinsulin has been investigated by characterization of split analogs containing single cleavages within or at the junctions of the C-domain (92). Internal cleavage (between residues 55 and 56 in human proinsulin) enhances receptor binding by only 2-fold, indicating that protein topology is not in itself the predominant factor in the low activity of proinsulin. Although cleavage at the BC and CA junctions augments binding by 5- and 15-fold, respectively, such analogs still exhibit relative activities of <25% (92). Together, these data suggest that the binding of proinsulin is impaired by long peptides tethered to the C terminus of the B-chain and N terminus of the A-chain, in accord with the constrained structure of the IR ectodomain (91).
SCIs in which the C-domain is either deleted or replaced by a short linker (<4 residues; “mini-proinsulins”) have found broad application in the recombinant expression of insulin precursors in yeast (13, 30, 84). The tightly constrained mini-proinsulins are essentially without biological activity (14, 15, 17). A prototype contains a direct peptide bond between LysB29 and GlyA1 with deletion of ThrB30. Although its affinity for the IR is reduced by >104-fold, the structure of this analog is essentially identical to that of wild-type insulin either as a zinc hexamer (15) or as an engineered monomer in solution (17). Such a marked loss of activity is ascribed to impaired induced fit: the tight linkage between the A- and B-chains presumably prevents their reorganization in a hormone-receptor complex (6, 14, 22). A variety of evidence suggests that the C-terminal β-strand of the B-chain detaches upon receptor binding (11, 15, 19-21, 23, 25).14
SCIs containing C-domains of intermediate length can exhibit substantial biological activity. 7-Residue C-domains have been described with activities of 14% (linker AAAAAAK) (18) and 28% (linker GGGPGKR) (32). The first example was provided by replacement of the wild-type C-domain of human proinsulin by the non-homologous 12-residue C-domain of IGF-I (16). The relative affinity of this 63-residue analog for the membrane-bound IR is 113%, whereas its relative affinity for a soluble ectodomain is 55-94% (16). Because the C-domain of IGF-I (unlike that of proinsulin) may introduce favorable contacts with the IR (93, 94), we propose that the high activity of this chimeric analog reflects the active engagement of its C-domain as an extension of the native receptor-binding surface. Such contacts presumably involve the cysteine-rich domain of the ectodomain and augment cross-binding to the Type I IGF receptor (19-28% relative to IGF-I), thus significantly reducing the specificity of the ligand (16). This proposal is supported by the observation that extending a 7-residue Ala-rich linker to 12 residues (C-domain AAAAAAAAAAAK) yields an SCI equal to the chimeric analog in length but of low activity (11%) (18). Given the potential contribution of foreshortened C-domains to affinity and specificity, it would be of future interest to select for optimal sequences by phage display as exemplified by studies of growth hormone (95).
Structure and Stability—The structure of SCI-57 is essentially identical to that of wild-type insulin. Its foreshortened C-domain is partially ordered and packs against a non-polar crevice between the A- and B-domains. The dibasic element extends the A1-A8 α-helix, and ProC4 in part packs against the side chain of ValA3, further sealing the classical hydrophobic core of insulin. These structural features are associated with increased thermodynamic stability as probed by guanidine denaturation studies. The extent of stabilization (ΔΔGu = 0.7 ± 0.2 kcal/mol relative to 2CA and 1.9 ± 0.2 kcal/mol relative to wild-type insulin) is unusual among conventional insulin analogs (96) but in accord with effects of bifunctional chemical cross-links between B29 and A1 (39).
Quantitative interpretation of protein denaturation experiments employs a two-state model for extrapolation of stabilities to zero denaturant concentration (57). Although this model represents an approximation, trends observed in comparative studies of related analogs are likely to be robust to the assumptions of the model. Comparison between single- and two-chain analogs may in principle be limited, however, by non-correspondence of respective denatured states. If more residual structure should be retained in a single-chain denatured state than in a two-chain denatured state, for example, this could lead to an underestimate of ΔΔGu (i.e. the SCI would be more stable than is indicated by the two-state model). This possibility seems unlikely in light of the difference in m values (Table 1), a parameter that is correlated with the extent of exposure of nonpolar surfaces upon unfolding (57). Whereas the m value of 2CA is 0.62 ± 0.02 kcal/mol/m (similar to those of AspB28-insulin (0.60 ± 0.01 kcal/mol/m) and wild-type insulin (0.58 ± 0.02 kcal/mol/m)), SCI-57 exhibits an m value of 0.79 ± 0.02 kcal/mol/m. This increase is consistent with a greater burial of non-polar surfaces in the native state, as predicted by the structure of the C-domain (see above) and decreased conformational fluctuations within the insulin moiety (see below). Greater retention of non-random structure in the single-chain denatured state would by contrast lead to a lower m value. Like SCI-57, the m value of human proinsulin is increased but to a lesser extent (0.71 ± 0.02 kcal/mol/m) (Table 1).
Because insulin functions as a monomer in the bloodstream, NMR studies of engineered monomers have attracted considerable attention. Although high resolution DG/simulated annealing models have been obtained (7, 9), the number of inter-residue NOEs under any one condition is generally smaller than is predicted based on back-calculation from such models or from the structures of crystallographic protomers (8, 77). This discrepancy has motivated the hypothesis that the insulin monomer (despite the high precision of NMR models) is in some respect “molten.” By contrast, the zinc-stabilized R6 insulin hexamer exhibits a rich density of NOEs (8, 10), presumably due to engagement of insulin within the confines of a well organized macromolecular assembly. Independent physical evidence for the damping of conformational fluctuations upon insulin assembly has been provided by laser Raman spectroscopy (97).
The present NMR studies provide particularly clear evidence for the anomalous dynamics of an engineered monomer. Comparison of the NOESY spectra of SCI-57 and 2CA demonstrates that whereas both proteins retain a native-like insulin fold, the single-chain analog exhibits more long-range NOEs within the insulin moiety than does 2CA. The subset of SCI-57-specific NOEs (see supplemental Table S2) are nonetheless consistent with interproton distances calculated from crystallographic protomers of wild-type insulin. We propose that like self-assembly, the foreshortened C-domain damps conformational fluctuations that would otherwise attenuate a subset of inter-residue NOEs. Because SCI-57 exhibits native activity, any barriers to inducted fit imposed by its greater conformational stability are presumably overcome by the favorable receptor contacts introduced by the HisA8 and AspB10 substitutions.
Therapeutic Implications—The complexity of insulin biosynthesis and of its regulation in the pancreatic β-cell has complicated the application of gene therapy to diabetes mellitus (98). The β-cell contains cell type-specific machinery for the processing of proinsulin and storage of insulin, including subtilisin-related prohormone convertases (99) and a granule-specific zinc import system. In an effort to bypass this β-cell-specific machinery, Lee et al. (32) described the glucose-regulated hepatic expression of an SCI. Introduced into the liver by an adeno-associated virus (100) and bearing a 7-residue linker (GGGPGKR), the SCI exhibited substantial but reduced affinity for the IR in the absence of proteolytic processing (32). Although the levels of expression in the bloodstream were not characterized and the extent of possible cleavage of the CA junction was not measured, the efficacy of this construct in glycemic control was demonstrated in a diabetic mouse model (32). Although the safety of such virus-based gene therapy in humans is uncertain, in principle, the use of an encoded SCI construct circumvents the complex pathway of insulin biosynthesis and storage in the β-cell.
Whereas gene therapy represents an experimental frontier of medical technology in the developed world, the developing world faces a challenge regarding the safe storage, delivery, and use of drugs and vaccines (101). This challenge complicates the use of temperature-sensitive insulin formulations in regions of Africa and Asia lacking consistent access to electricity and refrigeration (102), a challenge likely to be deepened by the pending epidemic of diabetes in the developing world (49). Insulin exhibits an increase in degradation rate of 10-fold or more for each 10 °C increment in temperature above 25 °C, and guidelines call for storage at temperatures <30 °C and preferably with refrigeration (103). At higher temperatures, insulin undergoes both chemical degradation (changes in covalent structure such as formation of isoaspartic acid, rearrangement of disulfide bridges, and formation of covalent polymers) and physical degradation (non-native aggregation and fibrillation) (50, 51). Preliminary studies of SCI-57 suggest that it is at least 60-fold more resistant than human insulin to degradation at 37 °C.
The development of ultrastable insulin formulations could aid in translating existing protocols for insulin replacement therapy to the Third World. Insulin is susceptible to diverse forms of chemical degradation at rates that are generally accelerated by conformational fluctuations and inversely correlated with thermodynamic stability (51). Although mechanisms of physical degradation are less well characterized (104), single-chain insulin analogs appear to exhibit an intrinsic resistance (relative to two-chain analogs) to non-native aggregation and fibrillation, proposed to reflect an incompatibility between chain topology and the structure of a cross-β-assembly (72). SCI-57 (with its striking combination of high activity, well organized structure, and augmented stability) may represent a class of insulin analogs whose pharmaceutical formulation could enhance the safety and efficacy of insulin replacement therapy. Accordingly, it would be of future interest to characterize the degradation pathways of SCI-57 and related analogs under conditions simulating those of patients in challenged regions of the developing world.
Supplementary Material
Acknowledgments
We thank P. De Meyts and Novo Nordisk Research Laboratories for the gift of human insulin; S. P. Yadav (Cleveland Clinic Foundation) for assistance with peptide synthesis; S. Wang for assistance with protein purification, Y.-M. Feng for providing PIP; Y. Yang for management of the NMR spectrometer; and P. Arvan, Y.-M. Feng, M. Liu, J. Whittaker, and Y. Yang for discussion and communication of results prior to publication. M. A. W. is grateful to P. De Meyts, G. G. Dodson, S. B. Kent, P. G. Katsoyannis, J. T. Potts, Jr., and D. F. Steiner for advice and encouragement.
The atomic coordinates and structure factors (code 2jzq) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant DK040949 (to M. A. W.). Native fragment ligation of SCI-57 was supported by United States Department of Energy Grant DE-FG02-07ER64501 (to S. B. Kent). This article is a contribution from the Cleveland Center for Structural Biology. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1-S5, Tables S1-S6, and additional references.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: IR, insulin receptor; SCI, single-chain insulin; PIP, porcine insulin precursor; RP-HPLC, reverse-phase high performance liquid chromatography; NOE, nuclear Overhauser enhancement; NOESY, NOE spectroscopy; TOCSY, total correlation spectroscopy; DG, distance geometry; IGF, insulin-like growth factor.
The insulin gene encodes a preproprotein containing (from the N to C terminus) a signal peptide, B-domain, C-domain, and A-domain. In proinsulin, the B-domain comprises residues 1-30; the C-domain, residues 31-66; and the A-domain, residues 66-86 (see Fig. 1A). In SCI-57, the respective B-, C-, and A-domains comprise residues 1-30, 31-36, and 37-57. For clarity and to facilitate comparison with previous studies of insulin, residues in SCI-57 are designated by position within the domain (e.g. CysB19, ProC4, and ValA3).
Evidence that redox-coupled refolding of SCI-57 led to its correct disulfide pairing is provided by its high activity and augmented stability, as mispairing generally leads to a significant reduction in activity and stability (105). Native pairing was explicitly verified by NMR observations of native-like disulfide-associated NOEs.
The crystal structure of des-ThrB30-SCI-50 has been determined as a native-like T3Rf3 zinc hexamer (where “Rf” is the frayed R-state in which B1 and B2 are not well ordered) (5); the NMR structure of a monomeric version of this analog (des-ThrB30-[AspB10, AspB28]SCI-50) has also been described (17). The crystal structure of an AspB28 analog of a 53-residue SCI (PIP) was determined as an R6 zinc hexamer containing seven m-cresol ligands/hexamer (33). Although a native-like hexamer was observed, distortion of the C-terminal B-chain β-strand occurs in one protomer of the component dimer associated with the AspB28 substitution and binding of m-cresol. The crystal structure of NαA1,NεB29-diaminosuberoyl insulin has also been determined (36). Although a native-like fold was observed within a T3Rf3 zinc hexamer, subtle distortions were observed in the A1-A8 α-helix interpreted as evidence of strain due to the cross-link. Guanidine denaturation studies of this analog nonetheless demonstrated augmented stability (ΔΔGu = 1.8 kcal/mol) (39).
The efficiency of insulin chain combination is generally limited by off-pathway aggregation and fibrillation of the B-chain, which competes with on-pathway interactions with the A-chain.
Low yields were unexpectedly encountered in attempted syntheses of [AspB10,AspB28,ProB29]insulin, suggesting that the substitution of ThrA8 by His rescued pairing of the variant B-chain. Because AspB10 enhances the efficiency of chain combination (29, 42, 61), it is possible that the AspB28,ProB29 element would by itself reduce the efficiency of chain combination.
J. Whittaker, B. Li, and M. A. Weiss, unpublished data.
The tractable 1H NMR spectrum of SCI-57 as a monomer in 20% deuterioacetic acid contrasts with the persistent self-association of des-ThrB30-SCI-50 in this co-solvent. The tightly constrained structure of the latter analog presumably maintains a well organized dimer interface, whereas the corresponding segments of SCI-57 and wild-type insulin are partially destabilized. The B29-A1 tether damps conformational fluctuations and so lowers the entropic cost of dimerization (106).
The interior of the inverted V-shaped IR structure appears empty in the deposited coordinates (Protein Data Bank code 2dtg) but in fact contains both carbohydrate and poorly ordered segments of the insert domain (91).
In classical structures, the C-terminal B-chain β-strand covers IleA2 and ValA3. Analogs containing allo-IleA2 (in which the chirality of theβ-carbon is inverted) (21) or LeuA3 exhibit native structure but low activity (25, 107). Destabilization of the B-chain β-strand by substitution of PheB24 by d-Phe enhances activity (26).
References
- 1.Dodson, G., and Steiner, D. (1998) Curr. Opin. Struct. Biol. 8 189-194 [DOI] [PubMed] [Google Scholar]
- 2.Blundell, T. L., Cutfield, J. F., Cutfield, S. M., Dodson, E. J., Dodson, G. G., Hodgkin, D. C., Mercola, D. A., and Vijayan, M. (1971) Nature 231 506-511 [DOI] [PubMed] [Google Scholar]
- 3.Smith, G. D., Swenson, D. C., Dodson, E. J., Dodson, G. G., and Reynolds, C. D. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 7093-7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, E. N., Blundell, T. L., Cutfield, J. F., Cutfield, S. M., Dodson, E. J., Dodson, G. G., Hodgkin, D. M., Hubbard, R. E., Isaacs, N. W., and Reynolds, C. D. (1988) Philos. Trans. R. Soc. Lond. B Biol. Sci. 319 369-456 [DOI] [PubMed] [Google Scholar]
- 5.Derewenda, U., Derewenda, Z., Dodson, E. J., Dodson, G. G., Reynolds, C. D., Smith, G. D., Sparks, C., and Swenson, D. (1989) Nature 338 594-596 [DOI] [PubMed] [Google Scholar]
- 6.Hua, Q.-x., Shoelson, S. E., Kochoyan, M., and Weiss, M. A. (1991) Nature 354 238-241 [DOI] [PubMed] [Google Scholar]
- 7.Hua, Q.-x., Hu, S. Q., Frank, B. H., Jia, W., Chu, Y. C., Wang, S. H., Burke, G. T., Katsoyannis, P. G., and Weiss, M. A. (1996) J. Mol. Biol. 264 390-403 [DOI] [PubMed] [Google Scholar]
- 8.Jacoby, E., Hua, Q.-x., Stern, A. S., Frank, B. H., and Weiss, M. A. (1996) J. Mol. Biol. 258 136-157 [DOI] [PubMed] [Google Scholar]
- 9.Olsen, H. B., Ludvigsen, S., and Kaarsholm, N. C. (1996) Biochemistry 35 8836-8845 [DOI] [PubMed] [Google Scholar]
- 10.Chang, X., Jorgensen, A. M., Bardrum, P., and Led, J. J. (1997) Biochemistry 36 9409-9422 [DOI] [PubMed] [Google Scholar]
- 11.Hua, Q.-x., Nakagawa, S. H., Hu, S. Q., Jia, W., and Weiss, M. A. (2006) J. Biol. Chem. 281 24900-24909 [DOI] [PubMed] [Google Scholar]
- 12.De Meyts, P., and Whittaker, J. (2002) Nat. Rev. Drug Discov. 1 769-783 [DOI] [PubMed] [Google Scholar]
- 13.Markussen, J., Jorgensen, K. H., Sorensen, A. R., and Thim, L. (1985) Int. J. Pept. Protein Res. 26 70-77 [PubMed] [Google Scholar]
- 14.Kobayashi, M., Sasaoka, T., Sugibayashi, M., Iwanishi, M., and Shigeta, Y. (1989) Diabetes Res. Clin. Pract. 7 25-28 [DOI] [PubMed] [Google Scholar]
- 15.Derewenda, U., Derewenda, Z., Dodson, E. J., Dodson, G. G., Bing, X., and Markussen, J. (1991) J. Mol. Biol. 220 425-433 [DOI] [PubMed] [Google Scholar]
- 16.Kristensen, C., Andersen, A. S., Hach, M., Wiberg, F. C., Schaffer, L., and Kjeldsen, T. (1995) Biochem. J. 305 981-986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua, Q.-x., Hu, S. Q., Jia, W., Chu, Y. C., Burke, G. T., Wang, S. H., Wang, R. Y., Katsoyannis, P. G., and Weiss, M. A. (1998) J. Mol. Biol. 277 103-118 [DOI] [PubMed] [Google Scholar]
- 18.Huang, Y., Liang, Z., and Feng, Y.-M. (2001) Sci. China 44 593-600 [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa, S. H., and Tager, H. S. (1989) J. Biol. Chem. 264 272-279 [PubMed] [Google Scholar]
- 20.Mirmira, R. G., and Tager, H. S. (1989) J. Biol. Chem. 264 6349-6354 [PubMed] [Google Scholar]
- 21.Xu, B., Hua, Q.-x., Nakagawa, S. H., Jia, W., Chu, Y. C., Katsoyannis, P. G., and Weiss, M. A. (2002) J. Mol. Biol. 316 435-441 [DOI] [PubMed] [Google Scholar]
- 22.Ludvigsen, S., Olsen, H. B., and Kaarsholm, N. C. (1998) J. Mol. Biol. 279 1-7 [DOI] [PubMed] [Google Scholar]
- 23.Huang, K., Chan, S. J., Hua, Q., Chu, Y. C., Wang, R., Klaproth, B., Jia, W., Whittaker, J., De Meyts, P., Nakagawa, S. H., Steiner, D. F., Katsoyannis, P. G., and Weiss, M. A. (2007) J. Biol. Chem. 282 35337-35349 [DOI] [PubMed] [Google Scholar]
- 24.Kitagawa, K., Ogawa, H., Burke, G. T., Chanley, J. D., and Katsoyannis, P. G. (1984) Biochemistry 23 1405-1413 [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa, S. H., and Tager, H. S. (1992) Biochemistry 31 3204-3214 [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, M., Ohgaku, S., Iwasaki, M., Maegawa, H., Shigeta, Y., and Inouye, K. (1982) Biochem. Biophys. Res. Commun. 107 329-336 [DOI] [PubMed] [Google Scholar]
- 27.Shoelson, S. E., Lu, Z. X., Parlautan, L., Lynch, C. S., and Weiss, M. A. (1992) Biochemistry 31 1757-1767 [DOI] [PubMed] [Google Scholar]
- 28.Markussen, J., Damgaard, U., Diers, I., Fiil, N., Hansen, M. T., Larsen, P., Norris, K., and Schou, O. (1987) in Peptides (Theodoropoulos, D., ed) pp. 189-194, Walter de Gruyter & Co., Berlin
- 29.Nakagawa, S. H., Zhao, M., Hua, Q.-x., Hu, S. Q., Wan, Z. L., Jia, W., and Weiss, M. A. (2005) Biochemistry 44 4984-4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markussen, J. (1985) Int. J. Pept. Protein Res. 25 431-434 [DOI] [PubMed] [Google Scholar]
- 31.Chang, S. G., Kim, D. Y., Choi, K. D., Shin, J. M., and Shin, H. C. (1998) Biochem. J. 329 631-635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, H. C., Kim, S. J., Kim, K. S., Shin, H. C., and Yoon, J. W. (2000) Nature 408 483-488 [DOI] [PubMed] [Google Scholar]
- 33.Whittingham, J. L., Edwards, D. J., Antson, A. A., Clarkson, J. M., and Dodson, G. G. (1998) Biochemistry 37 11516-11523 [DOI] [PubMed] [Google Scholar]
- 34.Brandenburg, D., and Wollmer, A. (1973) Hoppe-Seyler's Z. Physiol. Chem. 354 613-627 [DOI] [PubMed] [Google Scholar]
- 35.Freychet, P., Brandenburg, D., and Wollmer, A. (1974) Diabetologia 10 1-5 [DOI] [PubMed] [Google Scholar]
- 36.Cutfield, J., Cutfield, S., Dodson, E., Dodson, G., Hodgkin, D., and Reynolds, C. (1981) Hoppe-Seyler's Z. Physiol. Chem. 362 755-761 [DOI] [PubMed] [Google Scholar]
- 37.Gliemann, J., and Gammeltoft, S. (1974) Diabetologia 10 105-113 [DOI] [PubMed] [Google Scholar]
- 38.Cosmatos, A., Cheng, K., Okada, Y., and Katsoyannis, P. G. (1978) J. Biol. Chem. 253 6586-6590 [PubMed] [Google Scholar]
- 39.Brems, D. N., Brown, P. L., Nakagawa, S. H., and Tager, H. S. (1991) J. Biol. Chem. 266 1611-1615 [PubMed] [Google Scholar]
- 40.Kohn, W. D., Micanovic, R., Myers, S. L., Vick, A. M., Kahl, S. D., Zhang, L., Strifler, B. A., Li, S., Shang, J., Beals, J. M., Mayer, J. P., and DiMarchi, R. D. (2007) Peptides (N. Y.) 28 935-948 [DOI] [PubMed] [Google Scholar]
- 41.Brange, J., Ribel, U., Hansen, J. F., Dodson, G., Hansen, M. T., Havelund, S., Melberg, S. G., Norris, F., Norris, K., and Snel, L. (1988) Nature 333 679-682 [DOI] [PubMed] [Google Scholar]
- 42.Burke, G. T., Hu, S. Q., Ohta, N., Schwartz, G. P., Zong, L., and Katsoyannis, P. G. (1990) Biochem. Biophys. Res. Commun. 173 982-987 [DOI] [PubMed] [Google Scholar]
- 43.Brems, D. N., Alter, L. A., Beckage, M. J., Chance, R. E., DiMarchi, R. D., Green, L. K., Long, H. B., Pekar, A. H., Shields, J. E., and Frank, B. H. (1992) Protein Eng. 5 527-533 [DOI] [PubMed] [Google Scholar]
- 44.Brems, D. N., Brown, P. L., Bryant, C., Chance, R. E., Green, L. K., Long, H. B., Miller, A. A., Millican, R., Shields, J. E., and Frank, B. H. (1992) Protein Eng. 5 519-525 [DOI] [PubMed] [Google Scholar]
- 45.Kaarsholm, N. C., Norris, K., Jorgensen, R. J., Mikkelsen, J., Ludvigsen, S., Olsen, O. H., Sorensen, A. R., and Havelund, S. (1993) Biochemistry 32 10773-10778 [DOI] [PubMed] [Google Scholar]
- 46.Dawson, P. E., and Kent, S. B. H. (2000) Annu. Rev. Biochem. 69 923-960 [DOI] [PubMed] [Google Scholar]
- 47.Xu, B., Hu, S. Q., Chu, Y. C., Wang, S., Wang, R. Y., Nakagawa, S. H., Katsoyannis, P. G., and Weiss, M. A. (2004) Diabetes 53 1599-1602 [DOI] [PubMed] [Google Scholar]
- 48.Nakagawa, S. H., Hua, Q.-x., Hu, S. Q., Jia, W., Wang, S., Katsoyannis, P. G., and Weiss, M. A. (2006) J. Biol. Chem. 281 22386-22396 [DOI] [PubMed] [Google Scholar]
- 49.Lefebvre, P., and Pierson, A. (2004) World Hosp. Health Serv. 40 37-40, 42 [PubMed] [Google Scholar]
- 50.Pingel, M., and Volund, A. (1972) Diabetes 21 805-813 [DOI] [PubMed] [Google Scholar]
- 51.Brange, J., and Langkjoer, L. (1993) Pharm. Biotechnol. 5 315-350 [DOI] [PubMed] [Google Scholar]
- 52.Brange, J. (1987) Galenics of Insulin: The Physico-chemical and Pharmaceutical Aspects of Insulin and Insulin Preparations, Springer-Verlag, Berlin
- 53.Guthridge, S. L., and Miller, N. C. (1996) Aust. N. Z. J. Public Health 20 657-660 [DOI] [PubMed] [Google Scholar]
- 54.Chance, R. E., Hoffman, J. A., Kroeff, E. P., Johnson, M. G., Schirmer, W. E., and Bormer, W. W. (1981) in Proceedings of the Seventh American Peptide Symposium (Rich, D. H., and Gross, E., eds) pp. 721-728, Pierce Chemical Co., Rockford, IL
- 55.Hu, S. Q., Burke, G. T., Schwartz, G. P., Ferderigos, N., Ross, J. B., and Katsoyannis, P. G. (1993) Biochemistry 32 2631-2635 [DOI] [PubMed] [Google Scholar]
- 56.Schnolzer, M., Alewood, P., Jones, A., Alewood, D., and Kent, S. B. (1992) Int. J. Pept. Protein Res. 40 180-193 [DOI] [PubMed] [Google Scholar]
- 57.Sosnick, T. R., Fang, X., and Shelton, V. M. (2000) Methods Enzymol. 317 393-409 [DOI] [PubMed] [Google Scholar]
- 58.Hua, Q.-x., Jia, W., Frank, B. H., and Weiss, M. A. (1993) J. Mol. Biol. 230 387-394 [DOI] [PubMed] [Google Scholar]
- 59.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 60.Chakrabartty, A., and Baldwin, R. L. (1995) Adv. Protein Chem. 46 141-176 [PubMed] [Google Scholar]
- 61.Schwartz, G. P., Burke, G. T., and Katsoyannis, P. G. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 6408-6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang, S., Creagh, F. M., Peters, J. R., Brange, J., Volund, A., and Owens, D. R. (1991) Diabetes Care 14 571-577 [DOI] [PubMed] [Google Scholar]
- 63.Brooks, B., and Karplus, M. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 6571-6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kjeldsen, T., Ludvigsen, S., Diers, I., Balschmidt, P., Sorensen, A. R., and Kaarsholm, N. C. (2002) J. Biol. Chem. 277 18245-18248 [DOI] [PubMed] [Google Scholar]
- 65.Doig, A. J., and Baldwin, R. L. (1995) Protein Sci. 4 1325-1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nicholson, H., Anderson, D. E., Dao-pin, S., and Matthews, B. W. (1991) Biochemistry 30 9816-9828 [DOI] [PubMed] [Google Scholar]
- 67.Sancho, J., Serrano, L., and Fersht, A. R. (1992) Biochemistry 31 2253-2258 [DOI] [PubMed] [Google Scholar]
- 68.Bruch, M. D., Dhingra, M. M., and Gierasch, L. M. (1991) Proteins 10 130-139 [DOI] [PubMed] [Google Scholar]
- 69.Ciszak, E., Beals, J. M., Frank, B. H., Baker, J. C., Carter, N. D., and Smith, G. D. (1995) Structure (Lond.) 3 615-622 [DOI] [PubMed] [Google Scholar]
- 70.Hua, Q.-x., Chu, Y. C., Jia, W., Wang, R. Y., Katsoyannis, P. G., and Weiss, M. A. (2002) J. Biol. Chem. 277 43443-43453 [DOI] [PubMed] [Google Scholar]
- 71.Goldman, J., and Carpenter, F. H. (1974) Biochemistry 13 4566-4574 [DOI] [PubMed] [Google Scholar]
- 72.Huang, K., Maiti, N. C., Phillips, N. B., Carey, P. R., and Weiss, M. A. (2006) Biochemistry 45 10278-10293 [DOI] [PubMed] [Google Scholar]
- 73.Roy, M., Brader, M. L., Lee, R. W., Kaarsholm, N. C., Hansen, J. F., and Dunn, M. F. (1989) J. Biol. Chem. 264 19081-19085 [PubMed] [Google Scholar]
- 74.Weiss, M. A., Frank, B. H., Khait, I., Pekar, A., Heiney, R., Shoelson, S. E., and Neuringer, L. J. (1990) Biochemistry 29 8389-8401 [DOI] [PubMed] [Google Scholar]
- 75.Weiss, M. A., Hua, Q.-x., Lynch, C. S., Frank, B. H., and Shoelson, S. E. (1991) Biochemistry 30 7373-7389 [DOI] [PubMed] [Google Scholar]
- 76.Weiss, M. A., and Hoch, J. C. (1987) J. Magn. Reson. 72 324-333 [Google Scholar]
- 77.Hua, Q.-x., Ladbury, J. E., and Weiss, M. A. (1993) Biochemistry 32 1433-1442 [DOI] [PubMed] [Google Scholar]
- 78.Pullen, R. A., Lindsay, D. G., Wood, S. P., Tickle, I. J., Blundell, T. L., Wollmer, A., Krail, G., Brandenburg, D., Zahn, H., Gliemann, J., and Gammeltoft, S. (1976) Nature 259 369-373 [DOI] [PubMed] [Google Scholar]
- 79.Liang, D. C., Chang, W. R., and Wan, Z. L. (1994) Biophys. Chem. 50 63-71 [DOI] [PubMed] [Google Scholar]
- 80.Nakagawa, S. H., and Tager, H. S. (1986) J. Biol. Chem. 261 7332-7341 [PubMed] [Google Scholar]
- 81.Huang, K., Xu, B., Hu, S. Q., Chu, Y. C., Hua, Q.-x., Qu, Y., Li, B., Wang, S., Wang, R. Y., Nakagawa, S. H., Theede, A. M., Whittaker, J., De Meyts, P., Katsoyannis, P. G., and Weiss, M. A. (2004) J. Mol. Biol. 341 529-550 [DOI] [PubMed] [Google Scholar]
- 82.Xu, B., Hu, S. Q., Chu, Y. C., Huang, K., Nakagawa, S. H., Whittaker, J., Katsoyannis, P. G., and Weiss, M. A. (2004) Biochemistry 43 8356-8372 [DOI] [PubMed] [Google Scholar]
- 83.Weiss, M. A., Hua, Q.-x., Jia, W., Nakagawa, S. H., Chu, Y. C., and Katsoyannis, P. G. (2002) in Insulin and Related Proteins: Structure to Function and Pharmacology (Dieken, M. L., Federwisch, M., and De Meyts, P., eds) Kluwar Academic Publishers, Dordrecht, The Netherlands
- 84.Kjeldsen, T. (2000) Appl. Microbiol. Biotechnol. 54 277-286 [DOI] [PubMed] [Google Scholar]
- 85.Sohma, Y., Pentelute, B. L., Whittaker, J., Hua, Q.-x., Whittaker, L. J., Weiss, M. A., and Kent, S. B. H. (2007) Angew. Chem. Int. Ed. Engl. 47 1102-1106 [DOI] [PubMed] [Google Scholar]
- 86.Winter, J., Klappa, P., Freedman, R. B., Lilie, H., and Rudolph, R. (2002) J. Biol. Chem. 277 310-317 [DOI] [PubMed] [Google Scholar]
- 87.De Meyts, P. (1994) Diabetologia 37 S135-S148 [DOI] [PubMed] [Google Scholar]
- 88.Chan, S. J., Nakagawa, S., and Steiner, D. F. (2007) J. Biol. Chem. 282 13754-13758 [DOI] [PubMed] [Google Scholar]
- 89.Gauguin, L., Klaproth, B., Sajid, W., Andersen, A. S., McNeil, K. A., Forbes, B. E., and De Meyts, P. (2008) J. Biol. Chem. 283 2604-2613 [DOI] [PubMed] [Google Scholar]
- 90.Pekar, A. H., and Frank, B. H. (1972) Biochemistry 11 4013-4016 [DOI] [PubMed] [Google Scholar]
- 91.McKern, N. M., Lawrence, M. C., Streltsov, V. A., Lou, M. Z., Adams, T. E., Lovrecz, G. O., Elleman, T. C., Richards, K. M., Bentley, J. D., Pilling, P. A., Hoyne, P. A., Cartledge, K. A., Pham, T. M., Lewis, J. L., Sankovich, S. E., Stoichevska, V., Da Silva, E., Robinson, C. P., Frenkel, M. J., Sparrow, L. G., Fernley, R. T., Epa, V. C., and Ward, C. W. (2006) Nature 443 218-221 [DOI] [PubMed] [Google Scholar]
- 92.Peavy, D. E., Brunner, M. R., Duckworth, W. C., Hooker, C. S., and Frank, B. H. (1985) J. Biol. Chem. 260 13989-13994 [PubMed] [Google Scholar]
- 93.Cara, J. F., Mirmira, R. G., Nakagawa, S. H., and Tager, H. S. (1990) J. Biol. Chem. 265 17820-17825 [PubMed] [Google Scholar]
- 94.Denley, A., Bonython, E. R., Booker, G. W., Cosgrove, L. J., Forbes, B. E., Ward, C. W., and Wallace, J. C. (2004) Mol. Endocrinol. 18 2502-2512 [DOI] [PubMed] [Google Scholar]
- 95.Lowman, H. B., and Wells, J. A. (1993) J. Mol. Biol. 234 564-578 [DOI] [PubMed] [Google Scholar]
- 96.Weiss, M. A., Hua, Q.-x., Jia, W., Nakagawa, S. H., Chu, Y. C., Hu, S. Q., and Katsoyannis, P. G. (2001) J. Biol. Chem. 276 40018-40024 [DOI] [PubMed] [Google Scholar]
- 97.Dong, J., Wan, Z., Popov, M., Carey, P. R., and Weiss, M. A. (2003) J. Mol. Biol. 330 431-442 [DOI] [PubMed] [Google Scholar]
- 98.Selden, R. F., Skokiewicz, M. J., Russell, P. S., and Goodman, H. M. (1987) N. Eng. J. Med. 317 1067-1076 [DOI] [PubMed] [Google Scholar]
- 99.Steiner, D. F. (2000) J. Pediatr. Endocrinol. Metab. 13 229-239 [DOI] [PubMed] [Google Scholar]
- 100.Park, Y. M., Woo, S., Lee, G. T., Ko, J. Y., Lee, Y., Zhao, Z. S., Kim, H. J., Ahn, C. W., Cha, B. S., Kim, K. S., Park, C. W., and Lee, H. C. (2005) J. Gene Med. 7 621-629 [DOI] [PubMed] [Google Scholar]
- 101.Bott, R. F., and Oliveira, W. P. (2007) Drug Dev. Ind. Pharm. 33 393-401 [DOI] [PubMed] [Google Scholar]
- 102.102. Bassili, A., Omar, M., Tongnoni, G., and Egyptian-Italian Collaborative Group on Pediatric Chronic Diseases (2001) Diabetes Res. Clin. Pract. 53 187-199 [DOI] [PubMed] [Google Scholar]
- 103.Keller, K. J. (2003) South Dakota J. Med. 56 303-304 [PubMed] [Google Scholar]
- 104.Brange, J., Andersen, L., Laursen, E. D., Meyn, G., and Rasmussen, E. (1997) J. Pharm. Sci. 86 517-525 [DOI] [PubMed] [Google Scholar]
- 105.Hua, Q.-x., Jia, W., Frank, B. H., Phillips, N. F., and Weiss, M. A. (2002) Biochemistry 41 14700-14715 [DOI] [PubMed] [Google Scholar]
- 106.Tidor, B., and Karplus, M. (1994) J. Mol. Biol. 238 405-414 [DOI] [PubMed] [Google Scholar]
- 107.Wan, Z. L., Huang, K., Xu, B., Hu, S. Q., Wang, S., Chu, Y. C., Katsoyannis, P. G., and Weiss, M. A. (2005) Biochemistry 44 5000-5016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.