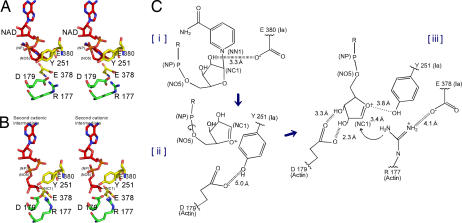
Fig. 4.
Mechanism of Sn1 ADP-ribosylation based on the strain-alleviation model. (A) Actin–Ia–NAD model. The structure is the same as that for actin–Ia–βTAD except that NAD is substituted for βTAD. (B) Second cationic intermediate. The N-ribose of ADP-ribose is rotated via NP-NO5 based on NAD. The torsion angles of three residues (Arg-177 and Asp-179) were manipulated to maintain appropriate bond lengths between atoms. (C) Schematic of the Sn1 mechanism of Ia: First, nicotinamide cleavage occurs via an Sn1 reaction induced by an NMN ring-like structure; second, the first oxocarbenium cation intermediate is formed with a strained conformation; third, the second cationic intermediate is induced by alleviation of the strained conformation by NP-NO5 rotation; fourth, actin Arg-177 of Ia nucleophilically attacks the NC1.