Fig. 3. Analysis of the association of SNAP-23 cysteine mutants with lipid rafts.
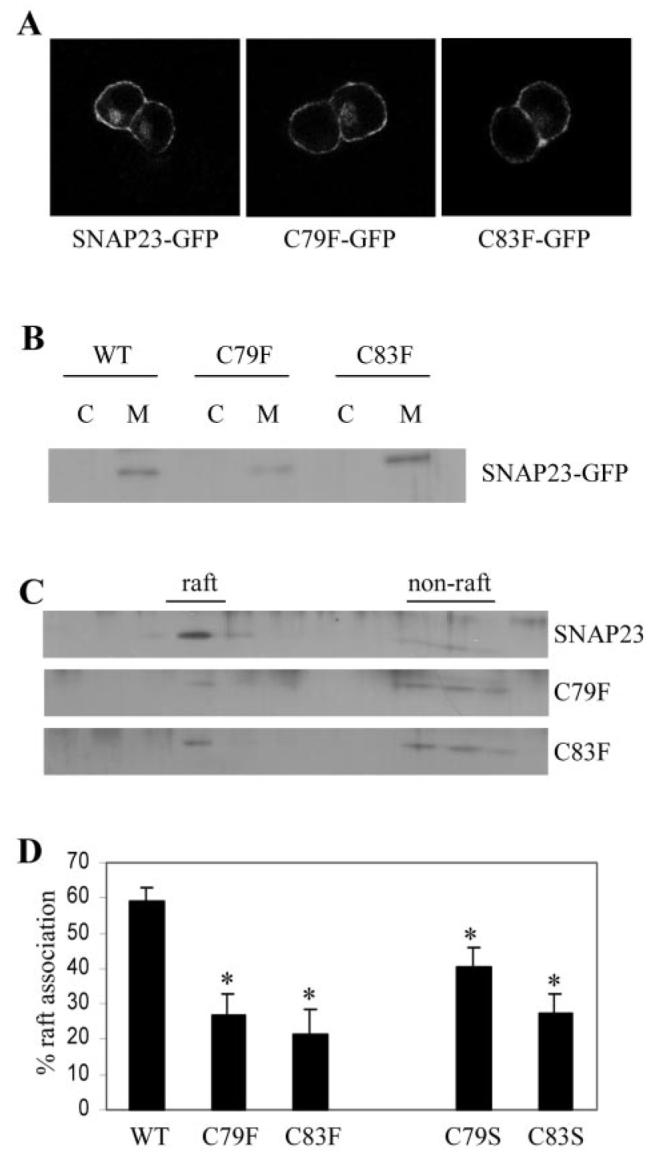
A, immunofluorescence analysis revealed that wild-type and mutant SNAP-23 proteins were strongly enriched at the cell surface. B, distribution of SNAP-23 and C79F and C83F mutants in cytosol (C) and membrane (M) fractions purified from PC12 cells. WT, wild type. C, membranes prepared from transfected PC12 cells were solubilized in Triton X-100 and fractionated on a discontinuous sucrose gradient as detailed under “Experimental Procedures.” Distribution of the GFP-tagged SNAP-23, C79F, and C83F proteins in the recovered gradient fractions was analyzed by immunoblotting using an antibody specific for GFP. D, averaged data from five separate experiments. *, p < 0.006 for C79F and C83F mutants when compared with wild-type SNAP-23. Also shown for comparison are normalized results for the raft association of C79S and C83S mutants, which displayed a marked reduction in raft association when compared with wild-type SNAP-23 (*, p < 0.02 for C79S and p < 0.0005 for C83S, n = 8)
