Figure 4.
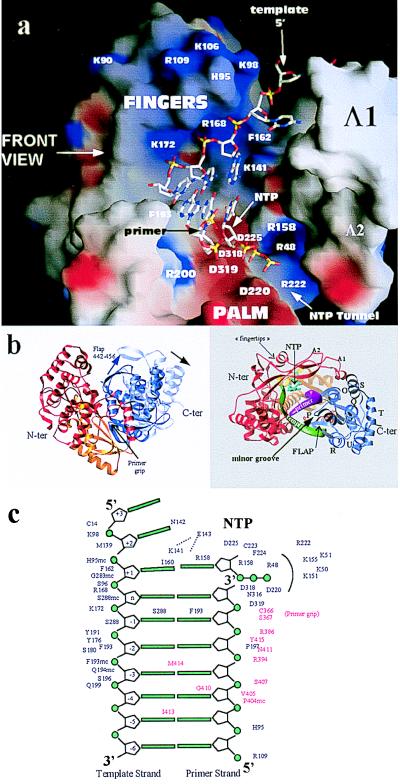
Interactions of the HCV RdRp with nucleic acid as inferred from the comparison with the structure of HIV-1 RT in a ternary complex with DNA and dTTP (26). (a) The template–primer duplex and dTTP molecule from the structure of the RT ternary complex superposed on a surface representation of the HCV polymerase colored according to electrostatic potential (as in Fig. 3 Upper). The view is a rotation of 90 degrees with respect to the front view of Figs. 1a and 3, looking from the thumb domain, which was removed for clarity. Some of the amino acids seen in the contacts are labeled. The transformation that superposes the palm and fingertips of the two enzymes has been applied to the nucleic acid coordinates for this figure. Five nucleotides of the template strand are shown, two of them base paired to the 3′ bases of the primer strand and a third to the incoming nucleotide, the triphosphate moiety of which points toward the tunnel mentioned in Fig. 3 and described in the text. (b) Proposed movement of the thumb on binding of RNA. The HCV polymerase is shown in a ribbon representation colored according to the different subdomains: fingers, red; palm, yellow; and thumb, blue. A rotation of ≈10 degrees (indicated by the black arrow in the left panel) about the connection between thumb and palm brings helix P in our structure into coincidence with helix H in HIV-1 RT, which tracks the minor groove of the A form DNA duplex. The resulting position of helix P in the complex is shown in the left panel, in a top view of the molecule where the template and primer backbone are displayed as green and magenta ribbons, respectively. The NTP molecule in the active site is colored cyan. The “flap” is a β-hairpin (strands 17 and 18), which has to move as well, as shown by the blue arrow in the left panel of the figure. (c) Diagram of the template–primer duplex in the catalytic site indicating the amino acid residues seen to contact the nucleic acid and nucleotide molecules after superposition to HIV-1 RT. These residues are very likely involved in binding RNA and nucleotide during polymerization. Residues behind an arc, around the triphosphate moiety of the dNTP molecule, are those lining the tunnel through which the NTP molecules are likely to access the active site of the enzyme. Residues indicated in red are those that come into contact after applying the rearrangement on the thumb shown in b.