TABLE 1.
Gene identification and expression profiles of selected genes
| Name
|
Description
|
NCBI accession no.
|
JGI v3.0 gene model name ID
|
Microarray heat mapa
|

|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Acid shock (assembly)
|
IBMX (disassembly)
|
||||||||||
| 0′ | 30′ | 60′ | 90′ | 0′ | 30′ | 60′ | 90′ | ||||
| Cellular processes and signaling | |||||||||||
| Cell motility | |||||||||||
| DHC1bc | Dynein heavy chain 1 α (PF9) | AJ243806.1 | estExt_GenewiseH_1.C_880001 | 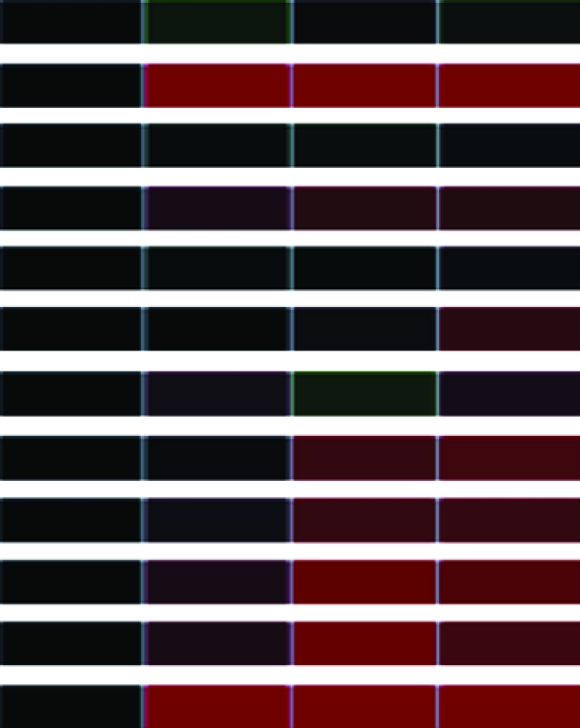 |
 |
||||||
| DLC7bc | LC7b, similar to the Roadblock/LC7 family | AY457969 | SAN_chlre3.20.7.3.11 | ||||||||
| IFT52bc | Intraflagellar transport 52 (BLD1) | AF420244 | estExt_fgenesh1_pm.C_150001 | ||||||||
| PF16c | Central pair-associated protein | U40057 | e_gwH.23.36.1 | ||||||||
| RSP3bc | Radial spoke protein 3 | X14549 | estExt_gwp_1W.C_260138 | ||||||||
| RSP4bc | Radial spoke protein 4 | M87526 | estExt_fgenesh1_pm.C_60005 | ||||||||
| RSP6bc | Radial spoke protein 6 | M87526 | estExt_GenewiseH_1.C_60055 | ||||||||
| TUA1bc | α-Tubulin 1 | M11447 | estExt_gwp_1H.C_140050 | ||||||||
| TUA2c | α-Tubulin 2 | M11448 | estExt_fgenesh2_kg.C_860004 | ||||||||
| TUB1bcd | β-Tubulin 1 | M10064 | estExt_gwp_1H.C_200099 | ||||||||
| TUB2bcd | β-Tubulin 2 | K03281 | estExt_gwp_1H.C_200090 | ||||||||
| VFL2bc | Centrin (caltractin) | X57973 | acegs_kg.scaffold_48000029 | ||||||||
| Post-translational modification, protein turnover, chaperones | |||||||||||
| HSP22A | Heat-shock protein 22A | X15053 | estExt_gwp_1W.C_60086 |  |
 |
||||||
| HSP22B | Heat-shock protein 22B | Chlre2_kg.scaffold_6000241 | |||||||||
| HSP22D | Heat-shock protein 22D | estExt_fgenesh2_pg.C_10198 | |||||||||
| HSP90A | Heat-shock protein 90A, localized to cytosol | estExt_gwp_1W.C_250014 | |||||||||
| HSP90C | Heat-shock protein 90C, localized to chloroplast | AY705371 | Chlre2_kg.scaffold_83000033 | ||||||||
| FAP277c | Flagellar-associated protein, protease inhibitor | SKA_Chlre2_kg.scaffold_28000189 | |||||||||
| LF4c | Long flagella protein | AY231293 | Chlre2_kg.scaffold_2000376 | ||||||||
| —e | Tyrosine-specific protein phosphatase | estExt_fgenesh2_pg.C_60262 | |||||||||
| —e | Unknown: AMPKbeta_GBD_like | fgenesh2_kg.C_scaffold_22000004 | |||||||||
| Signal transduction mechanisms | |||||||||||
| CRT2 | Calreticulin 2 | AJ000765 | estExt_GenewiseW_1.C_10142 | 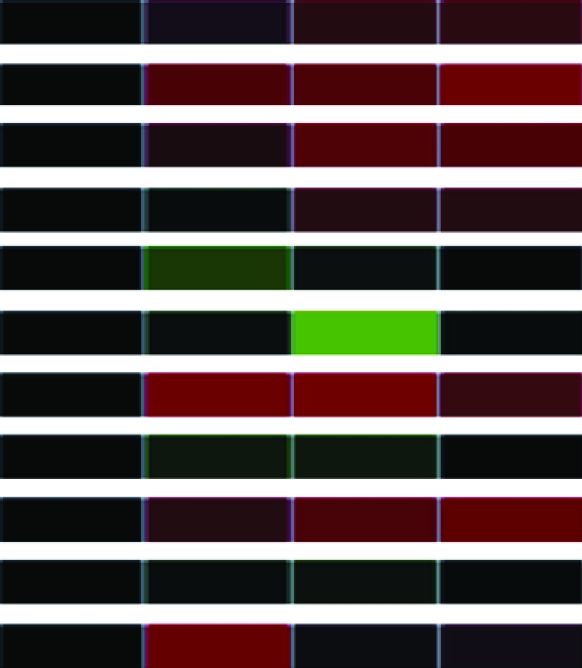 |
 |
||||||
| FAP12c | Flagellar-associated, lipase domain (pcf3-21) | estExt_fgenesh2_kg.C_240073 | |||||||||
| —e | G-protein-coupled seven transmembrane receptor | estExt_gwp_1H.C_310074 | |||||||||
| —de | GPR1/FUN34/yaaH, five transmembrane domains | e_gwH.13.239.1 | |||||||||
| —e | GPR1/FUN34/yaaH, six transmembrane domains | estExt_gwp_1W.C_130172 | |||||||||
| GUN4 | Similar to Gun4 (genomes uncoupled 4) | SKA_estExt_fgenesh2_pg.C_60106 | |||||||||
| MIPSd | Myo-inositol-1-phosphate synthase | estExt_fgenesh2_pg.C_90007 | |||||||||
| NDA3b | Putative mitochondrial NADH dehydrogenase | PIE_e_gwH.76.22.1 | |||||||||
| —de | Putative membrane protein, five transmembrane domains | estExt_fgenesh2_pg.C_20320 | |||||||||
| RACK1b | Receptor of activated protein kinase C 1 (pcf8-13) | e_gwH.38.29.1 | |||||||||
| —de | Thylakoid membrane protein, PGR5-like | estExt_gwp_1H.C_60062 | |||||||||
| Intracellular trafficking, secretion, vesicular transport | |||||||||||
| ARFA1ac | Small Arf-related GTPase | U27120 | estExt_fgenesh2_kg.C_110094 |  |
 |
||||||
| RABE1 | Small Rab-related GTPase | SAN_163210 | |||||||||
| Information storage and processing | |||||||||||
| Translation, ribosomal structure, and biogenesis | |||||||||||
| IF4Ac | Putative eukaryotic IF4A subunit (EIF41) | estExt_fgenesh2_pg.C_120244 | 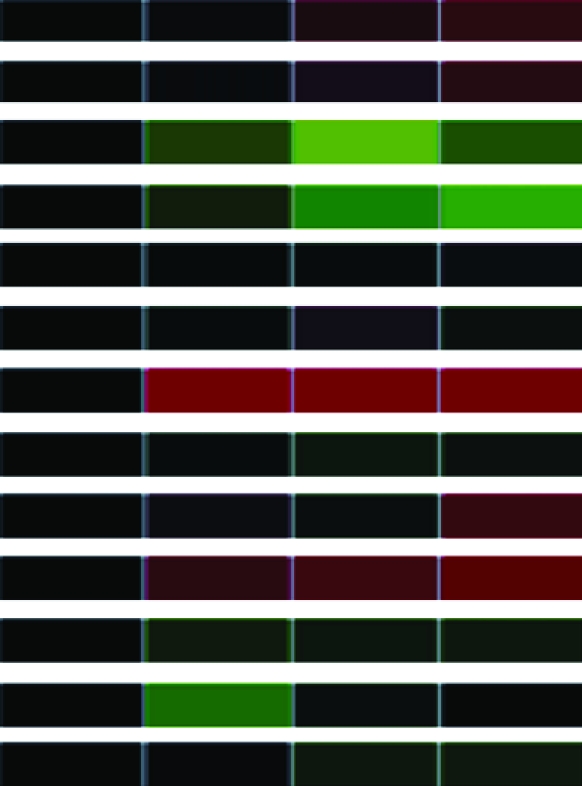 |
 |
|||||||
| MTT1 | Mitochondrial transcription termination factor like | estExt_fgenesh2_kg.C_390018 | |||||||||
| NOP56 | Nucleolar protein, component of C/D snoRNPs | estExt_gwp_1H.C_310180 | |||||||||
| PRPL7/L12 | Chloroplast ribosomal protein L7/L12 | e_gwH.2.436.1 | |||||||||
| RPL13 | Ribosomal protein L13 | estExt_gwp_1H.C_170118 | |||||||||
| RPL15 | Ribosomal protein L15 | estExt_GenewiseW_1.C_70201 | |||||||||
| RPL17 | Ribosomal protein L17 | estExt_GenewiseW_1.C_20197 | |||||||||
| RPL23a | Ribosomal protein L23a | fgenesh2_pg.C_scaffold_26000093 | |||||||||
| RPL27a | Ribosomal protein L27a | acegs_kg.scaffold_5000022 | |||||||||
| RPS16 | Ribosomal protein S16 | MAY_e_gwW.2.213.1 | |||||||||
| RPS19 | Ribosomal protein S19 | Chlre2_kg.scaffold_5000208 | |||||||||
| RPS3a | Ribosomal protein S3a | fgenesh2_pg.C_scaffold_2000130 | |||||||||
| —e | Small subunit ribosomal RNA-18S rDNA | M32703 | estExt_gwp_1H.C_4800003f | ||||||||
| Transcription | |||||||||||
| FAP280cd | Flagellar-associated, transcriptional coactivator-like | estExt_GenewiseW_1.C_50164 |  |
 |
|||||||
| —de | Putative zinc-finger protein, A20-like and AN1-like | OVA_e_gwH.25.97.1 | |||||||||
| —e | Putative zinc-finger domain, RING | Chlre2_kg.scaffold_11000031 | |||||||||
| Metabolism | |||||||||||
| Energy production and conversion | |||||||||||
| ANT1d | Adenine nucleotide translocator 1 | X65194 | JLM_estExt_gwp_1W.C_250131 |  |
 |
||||||
| COX2b | Cytochrome c oxidase subunit IIb (2b) | AF305540 | estExt_fgenesh2_pg.C_10741 | ||||||||
| CYB5_1 | Putative cytochrome b5 protein | estExt_gwp_1H.C_370036 | |||||||||
| GRX3 | Glutaredoxin 3, CGFS type, probably chloroplastic | estExt_fgenesh2_pg.C_810010 | |||||||||
| ICL1c | Isocitrate lyase | U18765 | estExt_fgenesh2_pg.C_260094 | ||||||||
| LHCA2 | Light-harvesting protein of photosystem I | Chlre2_kg.scaffold_11000154 | |||||||||
| LHCA5 | Light-harvesting protein of photosystem I | DQ370095 | estExt_fgenesh2_pg.C_80188 | ||||||||
| LHCB5 | Minor chlorophyll a-b-binding protein of PSII (CP26) | AB050007 | estExt_fgenesh2_kg.C_280035 | ||||||||
| LHCBM2d | Light-harvesting complex II chlorophyll a-b binding | AB051205 | estExt_fgenesh2_kg.C_200058 | ||||||||
| LHCBM3d | Light-harvesting complex II chlorophyll a-b binding M3 | AB051208 | estExt_fgenesh2_pg.C_940011 | ||||||||
| LHCBM9d | Light-harvesting complex II chlorophyll a-b binding | AF495472 | estExt_fgenesh2_kg.C_260046g | ||||||||
| PSAH | Subunit H of photosystem I (protein P28) | X15164 | estExt_fgenesh2_kg.C_100106 | ||||||||
| PSAI | Photosystem I subunit VIII, chloroplast-targeted | Chlre2_kg.scaffold_9000287 | |||||||||
| PTOX2 | Putative plastid terminal oxidase | AF494290 | SKA_estExt_fgenesh2_pg.C_90141 | ||||||||
| Amino acid transport and metabolism | |||||||||||
| —e | Asp/Glu racemase | estExt_fgenesh2_kg.C_10277 |  |
 |
|||||||
| HPD2 | 4-Hydroxyphenylpyruvate dioxygenase | AJ431261 | Chlre2_kg.scaffold_23000178 | ||||||||
| Carbohydrate transport and metabolism | |||||||||||
| PFK2 | Phosphofructokinase family (EC 2.7.1.11 or 2.7.1.90) | MHS_estExt_gwp_1W.C_200088 |  |
 |
|||||||
| RBCS2B | RuBisCO small subunit 2 | X04472 | estExt_GenewiseH_1.C_790027 | ||||||||
| UPTG1 | UDP-glucose:protein transglucosylase | estExt_fgenesh2_pg.C_20070 | |||||||||
| Coenzyme transport and metabolism | |||||||||||
| PDX1 | Pyridoxin biosynthesis protein | estExt_fgenesh2_pg.C_590022 | |||||||||
| Lipid transport and metabolism | |||||||||||
| SPT1 | Serine palmitoyltransferase | e_gwW.15.128.1 | |||||||||
| Ion transport and metabolism | |||||||||||
| ATS1 | ATP-sulfurylase | U57088 | HAL_estExt_fgenesh2_kg.C_190083 |  |
 |
||||||
| COP3d | Light-gated proton channel rhodopsin | AF508967 | Chlre2_kg.scaffold_43000011 | ||||||||
| Secondary metabolite biosynthesis, transport, and catabolism | |||||||||||
| HDS | 1-Hydroxy-2-methyl-2-(E)-butenyl4-diphosphate synthase | AY860817 | estExt_GenewiseH_1.C_110006 |  |
 |
||||||
| LAO1 | l-Amino acid oxidase | U78797 | estExt_fgenesh2_kg.C_200072 | ||||||||
| Poorly characterized | |||||||||||
| General function prediction only | |||||||||||
| CP12 | Small polypeptide associating with GAPDH and PRK | Chlre2_kg.scaffold_21000100 |  |
 |
|||||||
| FAP294c | Flagellar-associated coiled-coil protein | estExt_fgenesh2_pg.C_250096 | |||||||||
| FTTc | Fourteen-three-three, 14-3-3-like protein GF14 nu | X79445 | estExt_fgenesh2_kg.C_800003 | ||||||||
| LCI12 | Low-CO2-inducible protein | acegs_kg.scaffold_389000001 | |||||||||
| SMT1 | Similar to sterol-C24-methyltransferase | fgenesh2_kg.C_scaffold_11000008 | |||||||||
| ULP1 | Uncharacterized lumenal polypeptide | X96877 | estExt_fgenesh2_pg.C_10774 | ||||||||
| —e | Unknown, patatin domain | Chlre2_kg.scaffold_19000064 | |||||||||
| Function unknown | |||||||||||
| —e | Unknown | fgenesh2_pg.C_scaffold_60000034 |  |
 |
|||||||
| —e | Unknown | estExt_fgenesh2_kg.C_710007 | |||||||||
| —e | Unknown | estExt_fgenesh2_pg.C_230120 | |||||||||
| —e | Unknown | estExt_fgenesh2_pg.C_20470 | |||||||||
| —e | Unknown, three transmembrane domains | estExt_fgenesh2_kg.C_70077 | |||||||||
| —e | Unknown, eight transmembrane domains | Chlre2_kg.scaffold_12000168 | |||||||||
| —e | Unknown | OVA_Chlre2_kg.scaffold_21000142 | |||||||||
| Chlre2_kg.scaffold_8000021f | |||||||||||
| —e | Unknown, phosphoprotein-like | Chlre2_kg.scaffold_124000001 | |||||||||
Clones (118) sequenced and characterized by JGI and NCBI database searches were grouped into eukaryotic orthologous groups. Heat maps show relative mRNA abundance at each time point compared to that of untreated cells. Upregulation is represented in red, downregulation in green, and no change in black.
Heat maps generated using GeneSifter represent the collective data from three independent experiments.
PCR products were included on microarray for control purposes.
Found in the flagellar proteome according to Pazour et al. (2005) and/or JGI.
Genes represent more than one clone on microarray.
Genes not yet named.
Maps to multiple scaffolds on JGI.
Also maps on JGI to LHCBM4 (estExt_fgenesh2_pg.C_260118) and to LHCBM6 (estEXT_fgenesh2_kg.C_260057).
