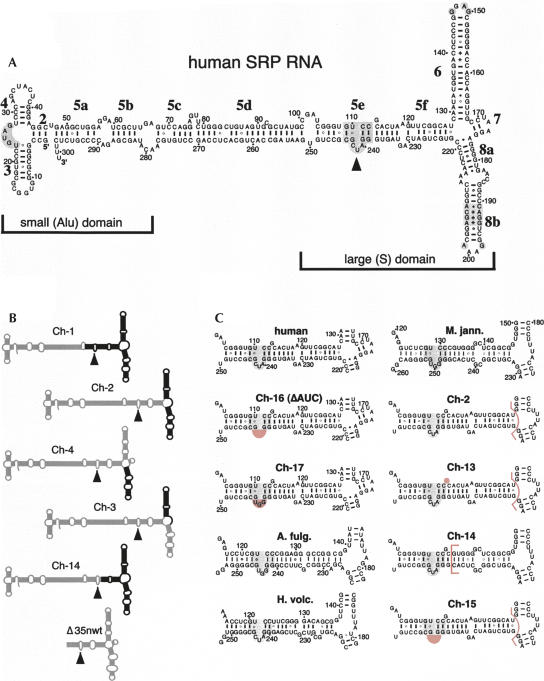
FIGURE 1.
Secondary structures of mutant SRP RNAs. (A) Secondary structure of human SRP RNA (hSR) with its small (Alu) and large (S) domains. Labeled are the 5′ and 3′ ends, the nucleotide residues in increments of 10, helices 2–8, and helical sections appended with letters a to f (Zwieb et al. 2005). Conserved regions are shaded gray and the 5e motif is marked by a solid arrowhead (Regalia et al. 2002). (B) Chimeric mutants Ch-1 to Ch-4 and Ch-14 are outlined with human SRP RNA sections in gray and Methanococcus jannaschii SRP RNA portions in black. (For details see Yin et al. 2001.) The Δ35nwt RNA corresponding to the large domain of the human SRP RNA is shown on the bottom. (C) Predicted secondary structures of the 5ef region in human SRP RNA (top, left column), Methanococcus jannaschii SRP RNA (M. jann., top, right column), the chimeric mutants Ch-2, Ch-13 to Ch-17, as well as the SRP RNAs of Archaeoglobus fulgidus (A. fulg.) and Haloferax volcanii (H. volc.). Ch-13 to Ch-15 are derivatives of Ch-2 as indicated by the pink bracket that separates the human from the M. jann. SRP RNA portions. Ch-16 (also named ΔAUC) and Ch-17 are alterations in the context of full-length human SRP RNA. The deletions of G113 in Ch-13, and of 240-AUC-242 (human SRP RNA numbering) in Ch-15 and Ch-16, as well as the AUC to GGU changes in Ch-17, are shaded pink. The 5e motif is shaded gray.