Abstract
Editing in trypanosomes involves the addition or deletion of uridines at specific sites to produce translatable mitochondrial mRNAs. RBP16 is an accessory factor from Trypanosoma brucei that affects mitochondrial RNA editing in vivo and also stimulates editing in vitro. We report here experiments aimed at elucidating the biochemical activities of RBP16 involved in modulating RNA editing. In vitro RNA annealing assays demonstrate that RBP16 significantly stimulates the annealing of gRNAs to cognate pre-mRNAs. In addition, RBP16 also facilitates hybridization of partially complementary RNAs unrelated to the editing process. The RNA annealing activity of RBP16 is independent of its high-affinity binding to gRNA oligo(U) tails, consistent with the previously reported in vitro editing stimulatory properties of the protein. In vivo studies expressing recombinant RBP16 in mutant Escherichia coli strains demonstrate that RBP16 is an RNA chaperone and that in addition to RNA annealing activity, it contains RNA unwinding activity. Our data suggest that the mechanism by which RBP16 facilitates RNA editing involves its capacity to modulate RNA secondary structure and promote gRNA/pre-mRNA annealing.
Keywords: RNA editing, trypanosomatids, RNA annealing, RNA chaperone, cold shock domain, Y-box protein
INTRODUCTION
Kinetoplastid protozoa, a group that includes human parasites Trypanosoma brucei, T. cruzi, and Leishmania spp., utilize U-insertion/deletion RNA editing, a unique and essential mechanism of gene regulation. Trypanosomes contain a single large mitochondrion whose biological functions are important for rapid response to environmental and life-cycle changes. In trypanosome mitochondria, most gene regulation occurs at post-transcriptional steps, including processing of polycistronic precursors, RNA stability, and RNA editing (Lukes et al. 2005). During kinetoplastid RNA editing, mitochondrial RNAs are modified by specific uridine insertion and deletion to form functional mRNAs, as directed by trans-acting guide RNAs (gRNAs). The sequence information for editing is transferred from mitochondrially encoded gRNAs that are complementary to the mature mRNA, through base-pairing interactions. Kinetoplastid RNA editing is catalyzed by an “enzyme cascade.” Editing is initiated by association of pre-mRNA and gRNA through formation of an anchor duplex between the gRNA 5′ end and mRNA region 3′ of the cognate editing domain, as well as interactions between the gRNA oligo(U) tail and pre-mRNA regions 5′ of the editing domain (Blum et al. 1990). Endonuclease cleavage of the mRNA is directed by gRNA/pre-mRNA base pairing. In the case of U insertion, uridines are added to the 3′ end of the 5′ cleavage product by a terminal uridylyl transferase (TUTase). In the case of U deletion, uridines are deleted by a U-specific exonuclease. The 5′- and 3′-end fragments are then religated by RNA ligases. The enzymatic activity is contained within several related multiprotein complexes, termed editosomes, that are comprised of between seven and 20 proteins (Rusche et al. 1997; Panigrahi et al. 2001, 2006; Aphasizhev et al. 2003a; Carnes et al. 2008). Editosomes contain all of the expected catalytic proteins, as well as several noncatalytic components important for protein–RNA or protein–protein interactions.
Several mitochondrial RNA-binding proteins have been identified in T. brucei that are not part of the editosome but may affect the accuracy and efficiency of RNA editing, including RBP16, MRP1/MRP2, TbRGG1, TbRGGm, and REAP1 (Koller et al. 1997; Madison-Antenucci et al. 1998; Vanhamme et al. 1998; Hayman and Read 1999; Blom et al. 2001; Panigrahi et al. 2007). RBP16 was initially identified in our laboratory as a mitochondrial protein that specifically binds gRNA. The affinity of RBP16 for gRNAs is mediated primarily through the 3′ oligo(U) tails (Hayman and Read 1999; Pelletier et al. 2000). RBP16 is composed of two domains, an N-terminal cold shock domain (CSD) and a C-terminal arginine/glycine rich (RGG) domain. In vitro, the CSD was shown to mediate high-affinity binding to the gRNA oligo(U) tail, while the RGG domain displays low-affinity, non-sequence-specific interactions with the gRNA (Miller and Read 2003). In vivo, through both immunoprecipitation and in organello cross-linking studies, RBP16 was shown to bind ∼30% of the total gRNAs (Hayman and Read 1999; Militello et al. 2000). Additionally, RBP16 associates with 9S and 12S rRNAs and certain mitochondrial mRNAs (Hayman and Read 1999; Goulah and Read 2007).
RNA interference (RNAi) studies revealed a role for RBP16 in both RNA editing and RNA stability in vivo (Pelletier and Read 2003). Disruption of RBP16 expression resulted in an ∼98% reduction in edited apocytochrome b (CYb) RNA and a corresponding increase in pre-edited CYb RNA, demonstrating a role for RBP16 in regulating RNA editing (Pelletier and Read 2003). A direct effect of RBP16 on RNA editing was subsequently confirmed by in vitro assays in which recombinant RBP16 significantly stimulated insertion editing at, or prior to, the step of pre-mRNA cleavage (Miller et al. 2006). RBP16 that is mutated within its CSD and is severely compromised for gRNA oligo(U) tail binding retained the ability to stimulate RNA editing, indicating that editing stimulation does not require high-affinity binding of the RBP16 CSD to the gRNA oligo(U) tail.
The CSD is a well conserved nucleic acid binding domain that is homologous to bacterial cold shock proteins (CSPs), and its presence defines RBP16 as a member of the eukaryotic Y-box protein family (Hayman and Read 1999). Y-box proteins consist of one or more CSDs, usually in conjunction with auxiliary domains. Some Y-box proteins, like the homologous CSPs, act as RNA chaperones and have been shown to contain RNA annealing and unwinding activity (Jiang et al. 1997; Skabkin et al. 2001; Nakaminami et al. 2006; Kim et al. 2007). RNA chaperone proteins are involved in numerous cellular processes including transcription, RNA processing, translation, and degradation (for review, see Cristofari and Darlix 2002).
In this paper, we characterize the biochemical activities of RBP16 and show that it is an RNA chaperone protein with RNA annealing and unwinding activity. Both the CSD and RGG domains of RBP16 contribute to the RNA annealing activity. RBP16 is capable of annealing both cognate gRNA/pre-mRNA pairs as well as complementary nonediting substrates. The gRNA/pre-mRNA annealing activity of RBP16 is independent of its high-affinity CSD-mediated binding to the gRNA oligo(U) tail, consistent with the previously reported RBP16 stimulation of in vitro RNA editing. These results suggest that the mechanism of RBP16 RNA editing stimulation involves modulation of RNA structure and gRNA/pre-mRNA annealing during the first step in RNA editing, prior to the catalytic steps of the reaction.
RESULTS
RBP16 stimulates the annealing of A6 gRNA to its cognate pre-mRNA
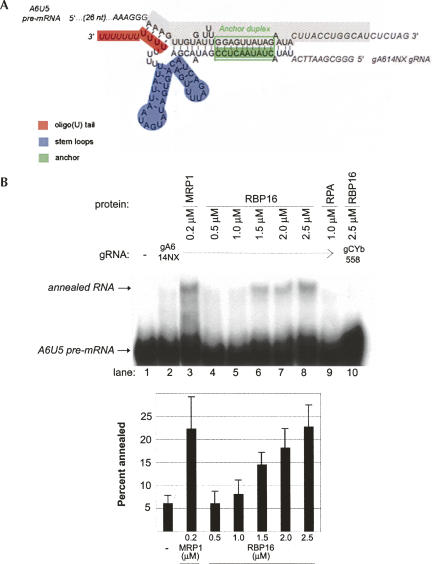
RNA editing initiates with formation of a gRNA/pre-mRNA hybrid, prior to pre-mRNA cleavage. We previously showed that RBP16 stimulates editing at, or prior to, the RNA cleavage step (Miller et al. 2006), suggesting that RBP16 might stimulate gRNA/pre-mRNA annealing. In addition, RBP16 is a member of the Y-box family of nucleic acid binding proteins (Hayman and Read 1999), another member of which, YB-1, has been shown to promote both the melting and annealing of nucleic acid strands (Skabkin et al. 2001). To determine if RBP16 is able to stimulate gRNA/pre-mRNA hybrid formation, we performed an RNA annealing assay using truncated ATPase 6 (A6) pre-mRNA molecules with edited sequence at editing site (ES) 1 (termed A6U5) and the cognate gA6[14]NX gRNA. We previously showed that editing at A6U5 ES2, as specified by gA6[14]NX, is stimulated by RBP16 in vitro (Miller et al. 2006). Formation of a gRNA/pre-mRNA hybrid results from annealing of the anchor duplex between the 3′-end of the pre-mRNA and 5′-end of the gRNA, as well as interactions between the gRNA 3′ oligo(U) tail and the pre-mRNA upstream purine-rich sequence (Fig. 1A; Koslowsky et al. 2004). Here, we assessed gRNA/pre-mRNA duplex formation by monitoring the reduced mobility of internally labeled A6U5 RNA bound to gA4[14]NX as compared with single-stranded A6U5 RNA in nondenaturing polyacrylamide gels (Muller et al. 2001). As shown in Figure 1B, in the absence of protein, addition of gA6[14]NX to radiolabeled A6U5 RNA resulted in only a small amount of input RNA forming a gRNA/pre-mRNA hybrid (Fig. 1B, cf. lanes 1 and 2). Addition of a known gRNA/pre-mRNA annealing factor, MRP1 (Muller et al. 2001; Aphasizhev et al. 2003b), stimulated RNA annealing approximately fourfold, as expected (Fig. 1B, lane 3). In the presence of increasing micromolar amounts of RBP16, we observed a concentration-dependent increase in cognate gRNA/pre-mRNA hybrid formation (Fig. 1B, lanes 4–8). Maximal RBP16-mediated stimulation of gRNA/pre-mRNA annealing was approximately fourfold using this RNA pair, similar to the extent of annealing stimulation by MRP1. Addition of a negative control protein, Replication Protein A (RPA), a single-stranded DNA-binding protein that has low affinity for RNA (Kim et al. 1992), had no effect on gRNA/pre-mRNA annealing (Fig. 1B, lane 9). To determine whether RBP16 annealing activity is specific for cognate gRNA/pre-mRNA pairs, we assayed the ability of RBP16 to stimulate duplex formation between A6U5 pre-mRNA and the noncognate CYb guide RNA, gCYb[558]. Even at the highest protein concentration, RBP16 did not stimulate formation of a duplex between the noncognate gRNA/pre-mRNA pair (Fig. 1B, lane 10). Together, these results demonstrate that RBP16 can stimulate annealing of a cognate gRNA/pre-mRNA pair.
FIGURE 1.
Recombinant RBP16 stimulates annealing of A6 pre-mRNA to cogate gRNA in vitro. (A) Diagram of the annealed A6U5 pre-mRNA (79 nt)/gA6[14]NX substrate pair, depicting gRNA domains and the gRNA/pre-mRNA anchor duplex (green box). The structure is based on the A6 gRNA/pre-mRNA secondary structure prediction from Leung and Koslowsky (1999). (Gray highlighted sequence) The truncated A6U5 41-nt pre-mRNA used in Fig. 3B. (B) Representative annealing assay with radiolabeled A6U5 pre-mRNA and gA6[14]NX gRNA at the indicated protein concentrations. (Lane 1) A6U5 pre-mRNA alone, (lane 2) A6U5 pre-mRNA and gA6[14]NX gRNA in the absence of protein, (lane 3) MRP1 positive control, (lanes 4–8) cognate gRNA/pre-mRNA pair in the presence of increasing RBP16, (lane 9) RPA negative control, (lane 10) reaction with RBP16 and a noncognate gRNA/pre-mRNA pair. Note that the amounts of RBP16 and MRP1 added are consistent with their known RNA binding affinities (Koller et al. 1997; Hayman and Read 1999). The bar graph represents the mean and standard error of the percent of the input pre-mRNA that is annealed to the gRNA from three separate experiments.
RBP16 RNA annealing is independent of high-affinity CSD–gRNA oligo(U) tail interaction
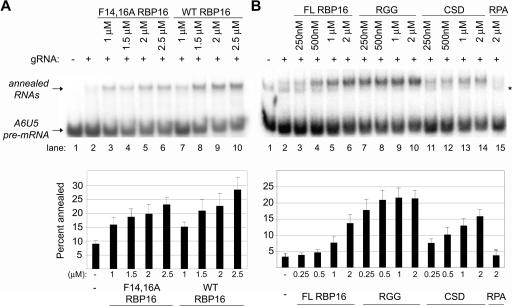
The RBP16 protein is composed of two distinct RNA-binding domains, an N-terminal CSD and a C-terminal RGG domain (Miller and Read 2003). The CSD mediates high-affinity binding to gRNA oligo(U) tails that is stable on mobility shift gels. The RGG domain displays a low-affinity interaction with the gRNA that is not stable in mobility shift gels but can be visualized by UV cross-linking. Therefore, the CSD contributes the majority of the gRNA binding avidity of RBP16. Mutation of conserved phenylalanines 14 and 16 of the RNP1 RNA-binding motif of the CSD dramatically reduces the gRNA binding ability of RBP16 (Miller et al. 2006). The F14,16A mutant is unable to form stable protein–RNA complexes in mobility shift assays; however, like the purified RGG domain, F14,16A RBP16 demonstrates some gRNA binding as evidenced by UV cross-linking. Importantly, F14,16A mutant RBP16 that is compromised in CSD-mediated RNA binding is capable of stimulating RNA editing in vitro to a similar extent as wild-type RBP16 (Miller et al. 2006). To determine whether RBP16-mediated editing stimulation and gRNA/pre-mRNA annealing stimulation share similar properties, we next asked whether the F14,16A RBP16 mutant is able to stimulate gRNA/pre-mRNA duplex formation. Annealing reactions were performed with both wild-type RBP16 and the CSD mutant F14,16A using the A6 RNA substrates described in Figure 1. As shown in Figure 2A, addition of increasing amounts of the F14,16A RBP16 resulted in a stimulation of RNA annealing (Fig. 2A, lanes 3–6), comparable to that observed with wild-type RBP16 (Fig. 2A, lanes 7–10). The ability of the mutant to stimulate annealing demonstrates that the CSD-mediated high-affinity interaction of RBP16 with RNA is not required for stimulation of gRNA/pre-mRNA duplex formation. These results suggest the high-affinity RNA-binding activity of RBP16 is not the mechanism by which RBP16 stimulates either RNA editing or RNA annealing, at least in in vitro assays with truncated pre-mRNA.
FIGURE 2.
Functions of RBP16 domains in RNA annealing. (A) RNA annealing reactions were performed as in Fig. 1 with radiolabeled A6U5 pre-mRNA and gA6[14]NX gRNA, using the indicated amounts of wild-type (WT) and F14,16A RBP16. (B) Representative RNA annealing reactions performed in the presence of the indicated amounts of full-length (FL) RBP16, RBP16 CSD, and RBP16 RGG domain proteins. Replication protein A (RPA) was used as a negative control. (*) An alternative A6U5 pre-mRNA secondary structure that is sometimes observed. The graphs represent the mean and standard error of three separate experiments.
Both the CSD and RGG domains of RBP16 are able to stimulate RNA annealing
Results from Figure 2A suggest that the RBP16 RGG domain may be critical in annealing stimulation, since CSD-mediated RNA interaction is not required. To directly determine the individual contributions of the CSD and RGG domains of RBP16 to dsRNA formation, both domains were purified as previously described (Miller and Read 2003) and used in annealing assays with the A6U5–gA6[14]NX RNA pair. RPA was used again as a negative control and showed no stimulation of dsRNA formation (Fig. 2B, lane 15). Interestingly, as shown in Figure 2B, addition of increasing amounts of either CSD (Fig. 2B, lanes 11–14) or RGG (Fig. 2B, lanes 7–10) was able to stimulate annealing at levels greater than or equal to the full-length RBP16 (Fig. 2B, lanes 3–6). These results demonstrate that both the CSD and the RGG domains possess RNA annealing activity, and that they can function individually. Because the RGG domain of RBP16 displays robust gRNA/pre-mRNA annealing activity, despite the fact that it lacks the high-affinity RNA binding conferred by the CSD, we conclude that RBP16 RNA annealing activity does not require high-affinity RNA binding. This is consistent with results in Figure 2A showing that the F14,16A RBP16 mutant retains RNA annealing activity even though RNA binding is seriously diminished.
The gRNA oligo(U) tail is not required for RBP16 stimulation of gRNA/pre-mRNA annealing
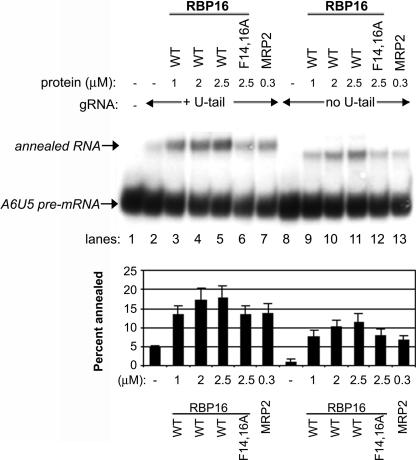
The CSD of RBP16 mediates high-affinity binding to the gRNA through the gRNA 3′ oligo(U) tail (Miller and Read 2003). However, Figure 2A shows that RBP16 harboring a CSD mutation that nearly abrogates gRNA binding still retains very significant gRNA/pre-mRNA annealing activity, indicating that CSD–oligo(U) tail interactions are not required for promotion of gRNA/pre-mRNA annealing. We cannot rule out, however, that the gRNA oligo(U) tail plays other important roles in gRNA/pre-mRNA duplex formation. For example, gRNA/pre-mRNA annealing involves interactions at the anchor duplex as well as through interaction of the gRNA oligo(U) tail with the pre-mRNA purine-rich sequence (Fig. 1A; Blum et al. 1990; Leung and Koslowsky 1999; Koslowsky et al. 2004), and the latter may contribute to overall gRNA/pre-mRNA duplex formation even if the U-tail is not specifically bound by annealing factors. To determine if the gRNA oligo(U) tail is required for RBP16 to stimulate dsRNA formation, RNA annealing experiments were performed with the A6U5 pre-mRNA used in Figures 1 and 2 and either the wild-type A6 gRNA that contains a 17-nucleotide (nt) oligo(U) tail (gA6[14]NX) or the same gRNA without an oligo(U) tail (gA6[14]NT). When equal molar amounts of either gRNA were added to the radiolabeled A6U5 pre-mRNA in the absence of protein, somewhat more duplex is formed in reactions with gA6[14]NX compared with gA6[14]NT (Fig. 3, cf. lanes 2 and 8). This is consistent with the U-tail stabilizing gRNA/pre-mRNA interaction, especially at low Mg2+ and RNA concentrations (Koslowsky et al. 2004). Recombinant T. brucei MRP2 was used as a positive control in these experiments and, as expected, stimulated annealing of gA6[14]NX to A6U5 pre-mRNA (Fig. 3, lane 7). Wild-type RBP16 also stimulated annealing of this RNA pair, as shown previously (Fig. 3, lanes 3–5). When we added increasing micromolar amounts of wild-type RBP16 to reactions containing A6U5 pre-mRNA and gA6[14]NT, which lacks an oligo(U) tail, we observed a significant stimulation of annealing in a range similar to that observed with the U-tailed gA6[14]NX (Fig. 3, cf. lanes 3–5 and 9–11). While the maximal percent annealed is slightly lower with gA6[14]NT compared with its U-tailed counterpart gA6[14]NX (Fig. 3, graph), the overall stimulation of gRNA/pre-mRNA annealing in the presence of gA6[14]NT (four- to fivefold) is slightly higher than that with gA6[14]NX (three- to fourfold), due to the lower duplex formation with gA6[14]NT in the absence of protein. We also observed that MRP2 was capable of substantial annealing stimulation with the gRNA lacking an oligo(U) tail (Fig. 3, lane 13). We next tested whether the CSD mutant F14,16A RBP16 was able to stimulate annealing with the U-less gRNA, gA6[14]NT, and found that it stimulated gRNA/pre-mRNA duplex formation to an extent similar to its stimulation in assays with U-tailed gRNA (Fig. 3, cf. lanes 6 and 12). These results demonstrate that wild-type and F14,16A RBP16, as well as MRP2, are able to stimulate annealing of A6 gRNA to pre-mRNA in the absence of the gRNA oligo(U) tail, and that this stimulation is comparable to reactions with U-tailed gRNAs. The results in Figure 3, as well as the results from Figure 2, demonstrate that the CSD-mediated high-affinity interaction of RBP16 with gRNA is not required for RBP16 to stimulate RNA duplex formation. We further conclude that the gRNA oligo(U) tail itself, and thus its interactions with the purine-rich region of the pre-mRNA upstream of the editing site, are not specifically required for RBP16 to stimulate gRNA/pre-mRNA annealing.
FIGURE 3.
RBP16 stimulation of gRNA/pre-mRNA annealing does not require the gRNA oligo(U) tail. Representative RNA annealing assays were performed as in Fig. 1 with radiolabeled A6U5 pre-mRNA and either A6 gRNA with a 17-nt oligo(U) tail (gA6[14]NX) or the same gRNA without an oligo(U) tail (gA6[14]NT). MRP2 was used as a positive control. (WT) Wild type. The graphs represent the mean and standard error of three separate experiments.
Intrinsic features of the RNAs limit the extent of annealing stimulation
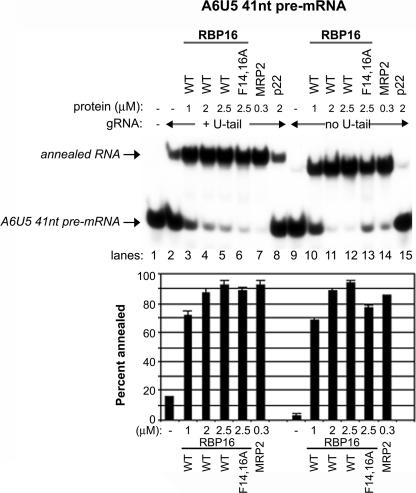
In Figures 1–3, the addition of RBP16, MRP1, or MRP2 stimulated gRNA/pre-mRNA hybrid formation, but this activity reached a plateau at 25%–30% of input pre-mRNA annealed. The inability of a large percentage of the A6 pre-mRNA to form a duplex with the gRNA may reflect limitations in the activities of annealing stimulatory proteins or in the RNAs themselves. In Figures 1–3, reactions were performed with the 79-nt-long A6U5 pre-mRNA. However, published experiments using a cross-linker at the 3′ end of a 10-nt-long U-tail of gA614 mapped a much smaller region of the A6-pre-mRNA interacting with the gA6[14] gRNA (Leung and Koslowsky 1999). To test the effect of pre-mRNA length on annealing activity, we prepared a truncated A6U5 pre-mRNA that we termed A6U5 41 nt. A6U5 41 nt has the same sequence as A6U5 pre-mRNA, but is truncated at the 5′ and 3′ ends (see Fig. 1A). Nevertheless, it contains all the sequence previously demonstrated to interact with the U(10)-gA6[14], including 13 nt upstream of the editing site and 33 nt downstream, encompassing the anchor sequence. Reactions in Figure 4 were performed as in previous experiments, but with the radiolabeled A6U5 41-nt pre-mRNA. In these experiments, we observed a much greater proportion of the input A6U5 41nt RNA annealed compared with that observed with the 79-nt A6U5 shorter (cf. Figs. 3 and 4). Addition of increasing amounts of RBP16, as well as the positive control MRP2, resulted in the majority of A6U5 41-nt pre-mRNA annealing to gA6[14]NX, with greater than fivefold stimulation even at relatively low protein concentrations. As with the 79 nt A6U5 shorter (Fig. 3), the oligo(U) tail of the gA6[14] was dispensable for annealing to A6U5 41 nt (Fig. 4, lanes 10–12). The mitochondrial protein, p22, which does not possess RNA-binding activity (Hayman et al. 2001), was used as a negative control and showed no stimulation of annealing (Fig. 4, lane 15). These results demonstrate that the limiting factor in gRNA/pre-mRNA annealing is not a property of the RBP16 or MRP2 proteins, but depends on the intrinsic characteristics of the RNAs being annealed.
FIGURE 4.
Features of the RNA limit annealing stimulation. Representative annealing assays were performed as in Fig. 1 except using a truncated A6 pre-mRNA (A6U5 41 nt). The sequence for A6U5 41 nt is highlighted in gray in Fig. 1A. MRP2 was used as a positive control and p22 protein was used as a negative control. (WT) Wild type. The graphs represent the mean and standard error of three separate experiments.
RBP16 stimulates annealing of partially complementary nonediting RNAs
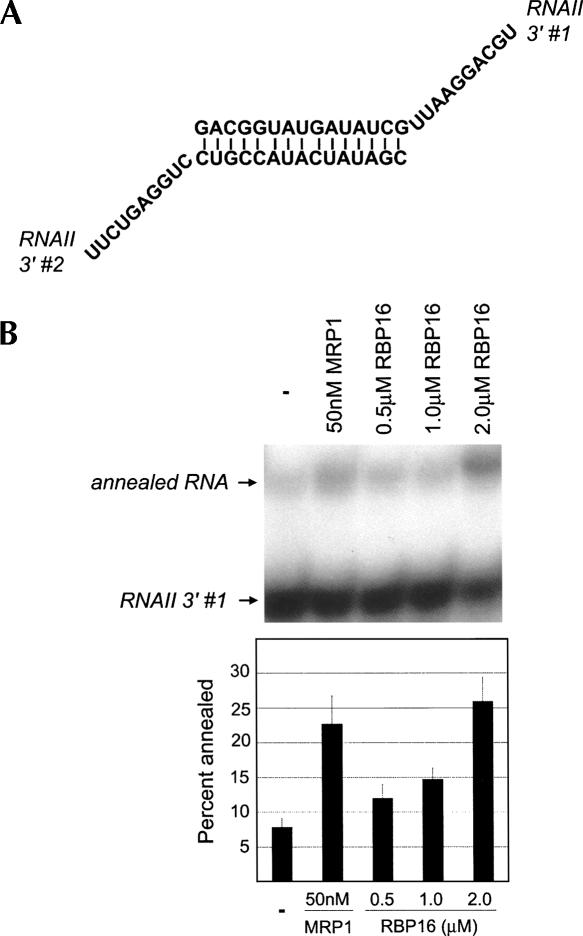
To this point, we have shown that RBP16, an editing accessory factor, stimulates annealing of gRNAs to pre-mRNAs. Next, we wanted to determine if the RNA annealing stimulation by RBP16 is specific to gRNA/pre-mRNA pairs or if the protein can stimulate annealing of nonediting RNAs. Previously, T. brucei MRP1 was shown to stimulate annealing of nonediting RNAs (Muller et al. 2001), and here we utilized one of these RNA pairs, RNAII 3′ #1 and #2, for the annealing assay. The synthetic RNAs contain a 15-base complementary sequence that is predicted to form a stem element and 10-base 3′ overhangs (Fig. 5A). Addition of the positive control, MRP1, or increasing amounts of RBP16 resulted in three- to fourfold stimulation of dsRNA formation (Fig. 5B). Thus, RBP16 has a general ability to anneal RNAs, and is not specific to editing substrates.
FIGURE 5.
RBP16 anneals nonediting RNA substrates. (A) Diagram of the annealed RNAII 3′ dsRNA (RNAII 3′ #1 and RNAII 3′ #2) containing a 15-bp duplex region with 10-nt 3′ overhangs (Muller et al. 2001). (B) Representative annealing assay with radiolabeled RNAII 3′ #1 and unlabeled RNAII 3′ #2 in the presence of the indicated protein concentrations. The graph represents the mean and standard error of three separate experiments.
RBP16 complements the activities of bacterial RNA chaperones
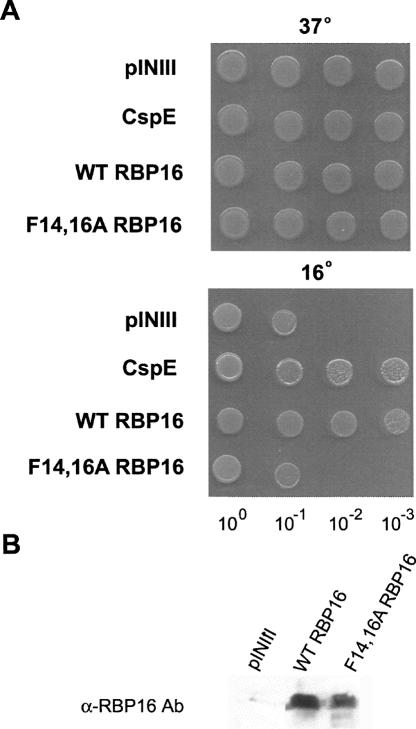
RNA annealing can be facilitated either through stabilization of the dsRNA, stabilization of the RNA in an annealing-competent conformation (matchmaker activity), or through RNA chaperone activity. RNA chaperones accelerate RNA folding by inducing changes in RNA secondary structure that resolve kinetically trapped RNA conformations or prevent their formation. Chaperones are no longer required once an RNA is properly folded or annealed (Herschlag 1995). RBP16 is a eukaryotic Y-box protein that contains an N-terminal CSD with homology to bacterial cold shock proteins. CSPs bind nucleic acids and function as RNA chaperones, and some CSPs are induced at high levels during cold stress (Graumann et al. 1997; Etchegaray and Inouye 1999; Cristofari and Darlix 2002). At lower temperatures, RNAs form very stable secondary structures, and the RNA chaperone activities of CSPs are thought to facilitate translation of RNAs at reduced temperatures by destabilizing RNA secondary structure. The Escherichia coli strain BX04 is a quadruple deletion mutant that lacks four CSPs (CspA, B, E, and G), and is thus cold sensitive. Numerous CSPs from bacteria and archaea, as well as eukaryotic Y-box proteins, have been shown to complement the cold sensitivity of BX04, presumably due to their ability to act as RNA chaperones (Nakaminami et al. 2006; Giaquinto et al. 2007; Kim et al. 2007). To determine whether RBP16 acts as an RNA chaperone in an in vivo context, we asked if it can complement cold sensitivity in the BX04 cells. To this end, wild-type RBP16 was cloned into the inducible pINIII vector and expressed in BX04 cells. All cells grew normally at the permissive temperature of 37°C, as expected (Fig. 6A, top). When cells were grown at the restrictive temperature (16°C), however, those cells containing the empty pINIII vector were not viable, and expression of E. coli CspE protein restored growth as previously reported (Phadtare et al. 2002; Fig. 6A, bottom). When RBP16 was expressed in BX04 cells, it clearly complemented the cold sensitivity, allowing growth at 16° at a level comparable to CspE (Fig. 6A, bottom). We next asked whether a functional CSD is required for the cold complementation property of RBP16 by assessing the complementation activity of F14,16A RBP16. Mutated RBP16 was not capable of restoring growth to the BX04 cells at the nonpermissive temperature, since cells expressing this protein grew similarly to cells containing empty vector (Fig. 6A, bottom). A Western blot using antibody against RBP16 confirmed comparable expression of wild-type and F14,16A RBP16 in BX04 cells (Fig. 6B). These data demonstrate that RBP16 is able to complement the activities of E. coli RNA chaperone proteins during the cold adaptation process, and that this activity requires a functional CSD. Based on the proposed functions of CSPs during cold stress, these results suggest that RBP16 acts as an RNA chaperone, destabilizing RNA secondary structure, in an in vivo context.
FIGURE 6.
RBP16 complements the low temperature sensitivity of an E. coli cspA, cspB, cspE, cspG quadruple mutant (BX04). (A) BX04 cells expressing empty vector (pINIII), wild-type (WT) RBP16, F14,16A RBP16, or the positive control CspE were grown overnight and diluted to an OD600 of 1. Tenfold serial dilutions were spotted out on LB plates with 50 μg/mL ampicillin and 0.2 mM IPTG and grown at 37°C (1 d) or 16°C (3–4 d). (B) Anti-RBP16 Western blot demonstrating equal expression of wild-type and F14,16A RBP16 in BX04 cells.
To directly ask whether the ability of RBP16 to rescue growth in BX04 cells during cold stress results from its capacity to modulate RNA structure, we performed an in vivo RNA chaperone assay that tests for transcription anti-termination activity (Bae et al. 2000; Nakaminami et al. 2006). A number of CSD-containing proteins have been characterized as RNA chaperones based on their anti-termination activity in this assay (Bae et al. 2000; Phadtare et al. 2003; Nakaminami et al. 2006). We utilized E. coli strain RL211, which contains a trpL terminator that forms a hairpin loop structure followed by the cat gene (Fig. 7A). Cells expressing proteins capable of unwinding the hairpin loop are able to grow in the presence of chloramphenicol. To test whether RBP16 displays anti-termination activity, the same pINIII constructs used to test cold stress complementation were transformed into RL211 cells. Cultures were grown in LB-ampicillin (+IPTG) and spotted onto LB-ampicillin (+IPTG) with or without chloramphenicol. Plates were incubated at 37°C for either 1 d (plates lacking chloramphenicol) or 4 d (plates containing chloramphenicol). As shown in Figure 7B, cells harboring empty vector (pINIII) were unable to grow in the presence of chloramphenicol, while those expressing bacterial CspE as a positive control displayed robust growth under the same conditions, as expected. Cells expressing wild-type RBP16 grew in the presence of chloramphenicol to a level comparable to CspE-expressing cells, demonstrating the anti-termination activity of RBP16. To test whether the CSD is required for anti-termination activity, we performed the same experiment with F14,16A RBP16. In contrast to wild-type RBP16, only background levels of growth were seen with cells expressing the F14,16A RBP16 mutant. Wild-type and F14,16A RBP16 were expressed at similar levels as judged by anti-RBP16 Western blot (Fig. 7C). The inability of F14,16A RBP16 to grow in the presence of chloramphenicol may result from a complete loss of RNA unwinding activity, or the activity may simply be decreased below the threshold required to support growth in this in vivo assay. Regardless, the data presented in Figure 7 demonstrate the importance of the N-terminal CSD of RBP16 in RNA unwinding, and therefore anti-termination, activity. Taken together, the results from Figures 6 and 7 support a role for RBP16 as an RNA chaperone and demonstrate that, in addition to RNA annealing activity, RBP16 also possesses RNA unwinding activity.
FIGURE 7.
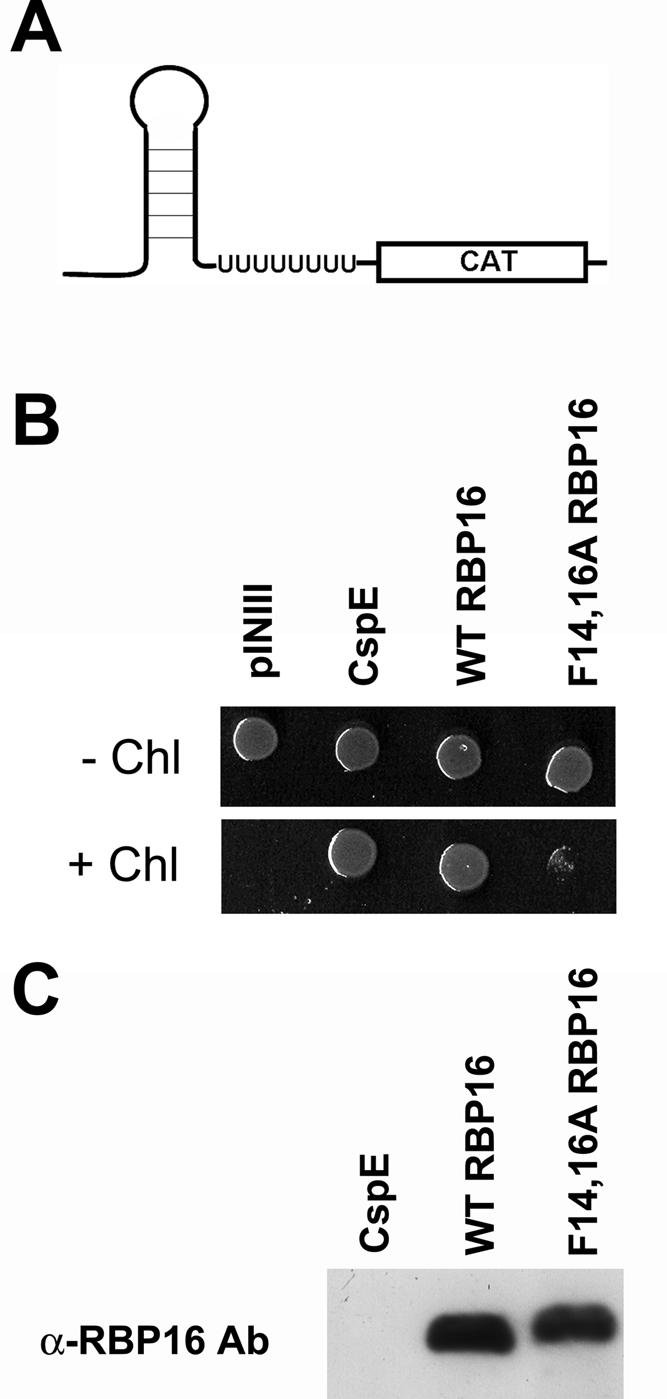
RBP16 demonstrates transcription anti-termination activity in E. coli strain RL211. (A) E. coli strain RL211 contains the trpL terminator that forms a hairpin loop upstream of a chloramphenicol (CAT) resistance gene. Expression of proteins capable of unwinding the RNA results in chloramphenicol resistance. (B) RL211 cells expressing empty vector (pINIII), wild-type (WT) RBP16, F14,16A RBP16, or the positive control CspE were grown in LB with 50 μg/mL ampicillin and 0.5 mM IPTG to an OD600 of 1 and spotted on LB plates containing 50 μg/mL ampicillin and 1 mM IPTG, with and without 30 μg/mL chloramphenicol. Cells were grown at 37°C for 1 d (− chloramphenicol) or 4 d (+ chloramphenicol). (C) Anti-RBP16 Western blot demonstrating equivalent expression of wild-type and F14,16A RBP16 in RL211 cells.
DISCUSSION
RBP16 was initially identified as a mitochondrial RNA-binding protein with high affinity for the oligo(U) tails of gRNAs. In addition to its in vivo association with gRNAs, it has been shown to associate with 9S and 12S rRNAs and a subset of mRNAs in T. brucei mitochondria (Hayman and Read 1999; Militello et al. 2000; Goulah and Read 2007). RBP16 was the first characterized RNA editing accessory factor, and both in vivo and in vitro assays demonstrate a role for RBP16 in stimulation of RNA editing (Pelletier and Read 2003; Miller et al. 2006). Two lines of evidence suggested that RBP16 might function in RNA editing through stimulation of RNA annealing. First, procyclic form trypanosomes disrupted for the MRP1/2 complex exhibit a phenotype very similar to that of RBP16 disruptants, and the MRP1/2 complex, as well as its component proteins, have been shown to facilitate gRNA/pre-mRNA annealing in vitro (Muller et al. 2001; Aphasizhev et al. 2003b; Vondruskova et al. 2005; Schumacher et al. 2006). Second, RBP16 is a Y-box protein that contains a CSD. Both eukaryotic CSD-containing proteins and the homologous prokaryotic CSPs have been shown to promote the melting and annealing of nucleic acid strands (Jiang et al. 1997; Skabkin et al. 2001; Nakaminami et al. 2006; Kim et al. 2007). Here, we report results characterizing the biochemical activities of the RBP16 protein. Using in vitro RNA annealing assays, we demonstrate that RBP16 stimulates the binding of gRNA to its cognate pre-mRNA to an extent similar to that of MRP1 and MRP2. The RNA annealing activity of RBP16 is independent of its high-affinity gRNA-binding capacity, and annealing does not require the gRNA oligo(U) tail. Indeed, RBP16 also stimulates annealing of complementary nonediting RNA pairs, an activity that it shares with MRP1/2 (Muller et al. 2001; Aphasizhev et al. 2003b). To address the mechanism by which RBP16 facilitates RNA annealing, we performed in vivo RNA chaperone assays in E. coli. We demonstrate that RBP16 is an RNA chaperone protein, and that in addition to its RNA annealing activity, RBP16 also facilitates RNA unwinding.
We performed a structure–function analysis of RBP16 to dissect the roles if its two constituent domains in RNA annealing. Both the N-terminal CSD and C-terminal RGG domains of RBP16 possess RNA-binding activity (Miller and Read 2003). The CSD mediates high-affinity, sequence-specific binding of RBP16 to gRNA through the oligo(U) tail, while the RGG domain mediates low-affinity nonspecific contacts with RNA (Miller and Read 2003). We performed in vitro RNA annealing assays in the presence of recombinant RBP16 CSD or RGG domain proteins. Interestingly, both individual domains displayed robust RNA annealing activity. Annealing activity of the RBP16 CSD is consistent with the demonstrated annealing activity of the eukaryotic CSD-containing proteins Prp24 and YB-1 (Raghunathan and Guthrie 1998; Skabkin et al. 2001). The RGG domain is highly basic, and stimulation of dsRNA formation by RGG is in agreement with the reported ability of proteins containing basic domains to promote RNA annealing (Buvoli et al. 1992; Hitti et al. 1998; Yang and Jankowsky 2005; Mayer et al. 2007). The weak nonspecific interaction of RGG with the RNA may in fact encourage its ability to promote RNA annealing by secondary structure destabilization. This inverse relationship between RNA binding and RNA annealing activity has previously been shown with the E. coli StpA protein (Mayer et al. 2007). The ability of the RGG domain to stimulate RNA annealing indicates that RBP16’s annealing stimulatory activity is independent of its high-affinity binding to the oligo(U) tail of the gRNA. This conclusion is supported by our results from in vitro RNA annealing assays performed with the RBP16 mutant, F14,16A, which contains mutations in the RNP1 RNA-binding motif of the CSD that abolish the high-affinity interaction with the gRNA oligo(U) tail (Miller et al. 2006). Even though the high-affinity gRNA binding is lost, F14,16A RBP16 retains the ability to stimulate RNA annealing. Thus, high-affinity RNA binding and RNA annealing are two distinct activities. It is likely that the annealing activity depends on non-sequence-specific interactions of RBP16 with the RNA. However, in the more complex in vivo milieu, it is possible that the specificity of RBP16 toward particular RNA targets is conferred by associated proteins.
One mechanism through which proteins act as annealing factors is through RNA chaperone activity. The presence of a CSD in RBP16 suggested that it may act in this manner. RNA chaperones are proteins that assist RNA molecules in correct folding by preventing or liberating the RNA from kinetically trapped and misfolded conformations (Herschlag 1995; Cristofari and Darlix 2002; Mayer et al. 2007). Chaperone proteins are defined by the fact they do not required a sequence-specific interaction, a specific RNA structure, or an energy source, and the protein is not required to be present after the RNA has been unfolded in order to maintain the correct RNA structure (Portman and Dreyfuss 1994; Herschlag 1995; Rein et al. 1998; Cristofari and Darlix 2002). Bacterial CSPs are important for cellular adaptation to cold stress, and their function at low temperature is directly related to their role as chaperone proteins (Jiang et al. 1997; Polissi et al. 2003). Eukaryotic proteins that contain CSDs have also been shown to act as RNA chaperones and can complement CSP function in vivo (Nakaminami et al. 2006; Kim et al. 2007). Similarly, RBP16 was able to rescue growth under cold stress in the E. coli cold-sensitive quadruple CSP deletion strain, BX04, demonstrating that RBP16 possesses RNA chaperone activity. RNA chaperones display diverse activities. Some RNA chaperones promote both RNA annealing and unwinding, while others may only be able to stimulate one of these reactions (Mayer et al. 2007). In this study, in vitro assays demonstrated that RBP16 possesses RNA annealing capacity. To examine the RNA unwinding capacity of RBP16, we performed an in vivo RNA chaperone assay that tests for transcription anti-termination activity (Bae et al. 2000; Phadtare et al. 2003; Nakaminami et al. 2006). Expression of RBP16, similar to bacterial CspE, facilitated transcription anti-termination and expression of a downstream chloramphenicol resistance gene, indicating that RBP16 has RNA unwinding activity in addition to RNA annealing activity. Thus, we can conclude that RBP16 is an RNA chaperone.
RBP16 was previously characterized as an RNA editing accessory factor in trypanosomes. RNA editing initiates with formation of the gRNA/pre-mRNA hybrid, prior to pre-mRNA cleavage. Evidence from in vitro RNA editing assays indicates that RBP16 stimulates editing at, or prior to, the cleavage of pre-mRNA (Miller et al. 2006), and RBP16 interacts with both gRNA and mRNA in vivo (Hayman and Read 1999; Goulah and Read 2007). In this paper, we have demonstrated that RBP16 has RNA annealing and unwinding activity. The ability of RBP16 to stimulate RNA annealing in vitro is consistent with a model in which the stimulation of RNA editing by RBP16 in vivo results from its capacity to facilitate gRNA annealing to pre-mRNA. The observation that RBP16 is not directly associated with editosomes suggests that RBP16 acts to promote editing prior to the catalytic steps of the process. The biochemical activities of the RBP16 F14,16A mutant also support this model. RBP16 F14,16A was previously shown to stimulate RNA editing in vitro to a similar extent as wild-type RBP16 (Miller et al. 2006). Therefore, the CSD–oligo(U) tail high-affinity interaction is not required for RBP16 editing stimulation in vitro. Likewise, we have shown here that the F14,16A mutant RBP16 is competent to stimulate RNA annealing. The fact that both the RNA annealing and editing stimulation of RBP16 are independent of binding to the gRNA oligo(U) tail supports a model in which the facilitation of gRNA/pre-mRNA annealing by RBP16 leads to stimulation of RNA editing. RBP16 might facilitate gRNA/pre-mRNA annealing by modulating gRNA structure, pre-mRNA structure, or both. In this regard, it is of interest that the pre-mRNA whose editing is most dramatically affected by RBP16 in vivo in PF T. brucei, CYb, has its anchor binding region positioned within a stable stem element (Koslowsky et al. 2004). Due to this stable pre-mRNA structure, gCYb[558] has a very low affinity for its cognate CYb pre-mRNA. Thus, it is likely that assistance in modulating CYb pre-mRNA structure is necessary for editing of this RNA to proceed, and RBP16 may perform such a function. Furthermore, we cannot rule out that in vivo RBP16 might also act as an RNA chaperone as editing proceeds subsequent to anchor duplex formation, counteracting kinetic roadblocks in gRNA and/or pre-mRNA structure.
It is important to note that although the F14,16A mutant RBP16 was able to stimulate RNA annealing in vitro, it was unable to support growth in the in vivo CSP complementation and anti-termination assays. These results imply that the F14,16A RBP16 mutant has either lost unwinding activity or has a reduced level of activity. This suggests several possibilities in regards to the role of unwinding in RNA editing. One is that RNA unwinding may not be involved in RNA editing either in vitro or in vivo. A second possibility is that RNA unwinding may be required in the complex in vivo environment, but not in a simpler in vitro setting. A final possibility is that the F14,16A mutant may have sufficient unwinding activity to support RNA editing in vivo and in vitro, although it may be insufficient to support E. coli growth in the assays used here. Whether the RNA unwinding activity of RBP16 contributes to its ability to stimulate editing in procyclic form trypanosomes will require additional studies with mutant proteins in T. brucei.
In summary, we have demonstrated that RBP16 is an RNA chaperone with both RNA annealing and RNA unwinding activities. The biochemical analysis of RBP16 undertaken in this study is a step forward in characterizing the role of RBP16 in stimulating RNA editing. In addition, RNA chaperone proteins are often ubiquitous and can perform multiple cellular functions (Cristofari and Darlix 2002; Kohno et al. 2003). RBP16 is abundant in procyclic form T. brucei and has been shown to function in RNA stabilization as well as RNA editing (Pelletier and Read 2003; Goulah et al. 2006). RNA stabilization may also involve modulation of RNA structure through RNA chaperone activity, possibly in mRNA 3′-untranslated regions. In addition to gRNA and mRNA, RBP16 also associates with rRNAs in trypanosome mitochondria, although the function of its rRNA binding capacity is unknown. Additional functions for RBP16 in the regulation of mitochondrial gene expression are yet to be identified, and the RNA chaperone activities of RBP16 may be important for these activities.
MATERIALS AND METHODS
Nucleic acid preparation
gA6[14]NX, gA6[14]NT, and gCYb[558] gRNAs were transcribed from plasmids as described previously (Read et al. 1994). A6U5 pre-mRNA (79 nt), GGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGGAAAGUUGUGAUUUUUGGAGUUAUAGAAUACUUACCUGGCAUCUCUAG, was synthesized from the A6 U5 shorter plasmid. RNAs were synthesized with the Ambion T7 Megascript kit. Transcripts were either internally labeled during transcription with [α 32P]-UTP or labeled after transcription at their 5′ ends with T4 polynucleotide kinase (Invitrogen) and [γ 32P]-ATP. The A6U5 41 nt, RNAII-3′ #1, and RNAII-3′ #2 RNAs were synthesized by Integrated DNA Technologies and labeled at their 5′ ends with T4 polynucleotide kinase. All RNAs were purified on a 6% acrylamide/7 M urea gel.
Protein expression and purification
Recombinant C-terminal His-tagged wild-type and F14,16A RBP16, as well as p22, were purified as previously described (Hayman et al. 2001). Recombinant C-terminal His-tagged CSD and RGG domains of RBP16 were purified as described (Miller and Read 2003). T. brucei MRP2 residues 30–224 (removing the N-terminal mitochondrial localization signal) (Schumacher et al. 2006) were cloned into the EcoRI–HindIII sites of pET42a (Novagen). Recombinant protein was expressed in the Rosetta E. coli strain (Novagen) and lysed by sonication in buffer A (50 mM Tris [pH 8.0], 250 mM NaCl, 5 mM EDTA, 10 mM DTT, and protease inhibitors) with lysozyme. Clarified lysate was incubated for 2 h with glutathione–agarose resin (Invitrogen), followed by extensive washes with buffer B (50 mM Tris [pH 8.7], 200 mM NaCl, 1 mM EDTA, 10 mM DTT). Purified protein was eluted with buffer B containing 10 mM reduced glutathione, pooled, and dialyzed against buffer B. Recombinant MRP1 (Ltp26) was generously provided by Ruslan Aphasizhev (Aphasizhev et al. 2003b). Recombinant human Replication Protein A (RPA) was a generous gift from Tom Melendy (Witebsky Center for Microbial Pathogenesis and Immunology) (Henricksen et al. 1994).
Annealing assays
Annealing reactions were performed similar to Muller et al. (2001) in 20 μL of buffer containing 4 mM HEPES-KOH (pH 7.5), 5 mM Tris-HCl (pH 7.5), 40 mM KCl, 2.2 mM MgCl2, 0.1 mM EDTA, 0.3 mM DTT, 2% glycerol, and 500 nM BSA. RNA substrates were incubated together for 2 min at 70°C, and cooled to room temperature for 15 min. Reactions contained ∼10 nM indicated radiolabeled RNA and 10 nM unlabeled RNA. The indicated protein amounts were added to the RNA mixture and incubated for 20 min at room temperature. Reactions were stopped by the addition of 40 μg of proteinase K, 0.1% SDS, 2.5 mM EDTA, and incubated for an additional 30 min. The reactions were diluted to a final volume of 40 μL with 4% glycerol, and 20 μL of each was loaded on an 8% native acrylamide gel in 100 mM Tris-HCl (pH 7.5) and 0.1 mM EDTA. The gel was run for 3 h at 10 W and analyzed using a Bio-Rad Quantity One PhosphorImager.
Plasmid construction
The pINIII and pINIII/cspE vectors were kindly provided by Dr. Sangita Phadtare (Robert Wood Johnson Medical School). The coding region of wild-type and F14,16A RBP16 were amplified from plasmids described previously (Miller et al. 2006) using primers Tb16K5′exp3, GCGAATTCCATATGAACAAGGGTAAGGTGATATCG and 3′expBamHI, CACGGATCCTCAAAAGTCATCGCTGAAGCTCTG. The PCR product was digested with NdeI and BamHI and cloned into the Nde/BamHI site of pINIII. These plasmids were designated pINIII-WTRBP16 and pINIII-F1416ARBP16.
In vivo chaperone assays
To test for the ability to complement cold sensitivity, empty pINIII expression vector or pINIII containing cspE, wild-type RBP16, or F14,16A RBP16 were transformed into E. coli BX04 mutant cells. BX04 cells lacking cold shock proteins CspA, B, E, and G and were kindly provided by Dr. Sangita Phadtare (Robert Wood Johnson Medical School) (Xia et al. 2001; Phadtare and Inouye 2004). Cells transformed with each vector were grown in LB media with ampicillin overnight and diluted into fresh media in the morning. When the cells had grown to an OD600 of 1, 10-fold serial dilutions were spotted on LB plates with 50 μg/mL ampicillin and 0.2 mM IPTG and cells were grown for 1 d at 37°C or 3–4 d at 16°C. To test anti-termination activity, the pINIII expression constructs were transformed into RL211 cells kindly provided by Dr. Robert Landick (University of Wisconsin) (Bae et al. 2000) and grown in LB with 50 μg/mL ampicillin and 0.5 mM IPTG to an OD600 of 1. Cultures were spotted on LB plates containing 50 μg/mL ampicillin and 1 mM IPTG, with and without 30 μg/mL chloramphenicol. Cells were grown for 1–4 d at 37°C. To analyze protein expression in pINIII constructs, 60 μL of each liquid culture was centrifuged prior to plating and cells resuspended in SDS-PAGE buffer. Half of each cell suspension was loaded on two 12.5% SDS-PAGE gels, one of which was Commassie stained and the other transferred to Hybond-P membrane (Amersham) at 50 V for 15 min. The membrane was then probed with a 1:2000 dilution of affinity-purified anti-RBP16 antibody (Hayman et al. 2001).
ACKNOWLEDGMENTS
We thank Dr. Ruslan Aphasizhev for generously providing recombinant MRP1 protein for use as a positive control in annealing assays and to Dr. Tom Melendy for providing RPA for use as a negative control. We also thank Dr. Sangita Phadtare for providing pINIII vectors, and Dr. Robert Landick for providing E. coli strain RL211. This work was supported by NIH grant RO1 AI061580 to L.K.R. M.L.A. was supported by American Heart Association Postdoctoral Fellowship # 0725813T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.982908.
REFERENCES
- Aphasizhev R., Aphasizheva I., Nelson R.E., Gao G., Simpson A.M., Kang X., Falick A.M., Sbicego S., Simpson L. Isolation of a U-insertion/-deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003a;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I., Nelson R.E., Simpson L. A 100-kDa complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003b;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W., Xia B., Inouye M., Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom D., Burg J., Breek C.K., Speijer D., Muijsers A.O., Benne R. Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21. Nucleic Acids Res. 2001;29:2950–2962. doi: 10.1093/nar/29.14.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B., Bakalara N., Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Buvoli M., Cobianchi F., Riva S. Interaction of hnRNP A1 with snRNPs and pre-mRNAs: Evidence for a possible role of A1 RNA annealing activity in the first steps of spliceosome assembly. Nucleic Acids Res. 1992;20:5017–5025. doi: 10.1093/nar/20.19.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J., Trotter J.R., Peltan A., Fleck M., Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell. Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G., Darlix J.L. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- Etchegaray J.P., Inouye M. CspA, CspB, and CspG, major cold shock proteins of Escherichia coli, are induced at low temperature under conditions that completely block protein synthesis. J. Bacteriol. 1999;181:1827–1830. doi: 10.1128/jb.181.6.1827-1830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaquinto L., Curmi P.M., Siddiqui K.S., Poljak A., DeLong E., DasSarma S., Cavicchioli R. Structure and function of cold shock proteins in archaea. J. Bacteriol. 2007;189:5738–5748. doi: 10.1128/JB.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulah C.C., Read L.K. Differential effects of arginine methylation on RBP16 mRNA binding, guide RNA (gRNA) binding, and gRNA-containing ribonucleoprotein complex (gRNP) formation. J. Biol. Chem. 2007;282:7181–7190. doi: 10.1074/jbc.M609485200. [DOI] [PubMed] [Google Scholar]
- Goulah C.C., Pelletier M., Read L.K. Arginine methylation regulates mitochondrial gene expression in Trypanosoma brucei through multiple effector proteins. RNA. 2006;12:1545–1555. doi: 10.1261/rna.90106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P., Wendrich T.M., Weber M.H., Schroder K., Marahiel M.A. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 1997;25:741–756. doi: 10.1046/j.1365-2958.1997.5121878.x. [DOI] [PubMed] [Google Scholar]
- Hayman M.L., Read L.K. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA-binding activity. J. Biol. Chem. 1999;274:12067–12074. doi: 10.1074/jbc.274.17.12067. [DOI] [PubMed] [Google Scholar]
- Hayman M.L., Miller M.M., Chandler D.M., Goulah C.C., Read L.K. The trypanosome homolog of human p32 interacts with RBP16 and stimulates its gRNA-binding activity. Nucleic Acids Res. 2001;29:5216–5225. doi: 10.1093/nar/29.24.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricksen L.A., Umbricht C.B., Wold M.S. Recombinant replication protein A: Expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Hitti E., Neunteufl A., Jantsch M.F. The double-stranded RNA-binding protein X1rbpa promotes RNA strand annealing. Nucleic Acids Res. 1998;26:4382–4388. doi: 10.1093/nar/26.19.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Hou Y., Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- Kim C., Snyder R.O., Wold M.S. Binding properties of replication protein A from human and yeast cells. Mol. Cell. Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Park S.J., Kwak K.J., Kim Y.O., Kim J.Y., Song J., Jang B., Jung C.H., Kang H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli . Nucleic Acids Res. 2007;35:506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K., Izumi H., Uchiumi T., Ashizuka M., Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Koller J., Muller U.F., Schmid B., Missel A., Kruft V., Stuart K., Goringer H.U. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 1997;272:3749–3757. doi: 10.1074/jbc.272.6.3749. [DOI] [PubMed] [Google Scholar]
- Koslowsky D.J., Reifur L., Yu L.E., Chen W. Evidence for U-tail stabilization of gRNA/mRNA interactions in kinetoplastid RNA editing. RNA Biol. 2004;1:28–34. doi: 10.4161/rna.1.1.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung S.S., Koslowsky D.J. Mapping contacts between gRNA and mRNA in trypanosome RNA editing. Nucleic Acids Res. 1999;27:778–787. doi: 10.1093/nar/27.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukes J., Hashimi H., Zikova A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci S., Sabatini R.S., Pollard V.W., Hajduk S.L. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): A novel editing complex protein with repetitive domains. EMBO J. 1998;17:6368–6376. doi: 10.1093/emboj/17.21.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer O., Rajkowitsch L., Lorenz C., Konrat R., Schroeder R. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 2007;35:1257–1269. doi: 10.1093/nar/gkl1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Militello K.T., Hayman M.L., Read L.K. Transcriptional and post-transcriptional in organello labelling of Trypanosoma brucei mitochondrial RNA. Int. J. Parasitol. 2000;30:643–647. doi: 10.1016/s0020-7519(00)00041-2. [DOI] [PubMed] [Google Scholar]
- Miller M.M., Read L.K. Trypanosoma brucei: Functions of RBP16 cold shock and RGG domains in macromolecular interactions. Exp. Parasitol. 2003;105:140–148. doi: 10.1016/j.exppara.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Miller M.M., Halbig K., Cruz-Reyes J., Read L.K. RBP16 stimulates trypanosome RNA editing in vitro at an early step in the editing reaction. RNA. 2006;12:1292–1303. doi: 10.1261/rna.2331506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U.F., Lambert L., Goringer H.U. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 2001;20:1394–1404. doi: 10.1093/emboj/20.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K., Karlson D.T., Imai R. Functional conservation of cold shock domains in bacteria and higher plants. Proc. Natl. Acad. Sci. 2006;103:10122–10127. doi: 10.1073/pnas.0603168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Gygi S.P., Ernst N.L., Igo R.P., Jr, Palazzo S.S., Schnaufer A., Weston D.S., Carmean N., Salavati R., Aebersold R., et al. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Ernst N.L., Domingo G.J., Fleck M., Salavati R., Stuart K.D. Compositionally and functionally distinct editosomes in Trypanosoma brucei . RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Zikova A., Dalley R.A., Acestor N., Ogata Y., Anupama A., Myler P.J., Stuart K.D. Mitochondrial complexes in Trypanosoma brucei: A novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics. 2007;3:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- Pelletier M., Read L.K. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei . RNA. 2003;9:457–468. doi: 10.1261/rna.2160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M., Miller M.M., Read L.K. RNA-binding properties of the mitochondrial Y-box protein RBP16. Nucleic Acids Res. 2000;28:1266–1275. doi: 10.1093/nar/28.5.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadtare S., Inouye M. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli . J. Bacteriol. 2004;186:7007–7014. doi: 10.1128/JB.186.20.7007-7014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadtare S., Inouye M., Severinov K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002;277:7239–7245. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- Phadtare S., Hwang J., Severinov K., Inouye M. CspB and CspL, thermostable cold-shock proteins from Thermotoga maritima . Genes Cells. 2003;8:801–810. doi: 10.1046/j.1365-2443.2003.00675.x. [DOI] [PubMed] [Google Scholar]
- Polissi A., De Laurentis W., Zangrossi S., Briani F., Longhi V., Pesole G., Deho G. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 2003;154:573–580. doi: 10.1016/S0923-2508(03)00167-0. [DOI] [PubMed] [Google Scholar]
- Portman D.S., Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13:213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan P.L., Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- Read L.K., Goringer H.U., Stuart K. Assembly of mitochondrial ribonucleoprotein complexes involves specific guide RNA (gRNA)-binding proteins and gRNA domains but does not require preedited mRNA. Mol. Cell. Biol. 1994;14:2629–2639. doi: 10.1128/mcb.14.4.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A., Henderson L.E., Levin J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: Significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- Rusche L.N., Cruz-Reyes J., Piller K.J., Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M.A., Karamooz E., Zikova A., Trantirek L., Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- Skabkin M.A., Evdokimova V., Thomas A.A., Ovchinnikov L.P. The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J. Biol. Chem. 2001;276:44841–44847. doi: 10.1074/jbc.M107581200. [DOI] [PubMed] [Google Scholar]
- Vanhamme L., Perez-Morga D., Marchal C., Speijer D., Lambert L., Geuskens M., Alexandre S., Ismaili N., Goringer U., Benne R., et al. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem. 1998;273:21825–21833. doi: 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- Vondruskova E., van den Burg J., Zikova A., Ernst N.L., Stuart K., Benne R., Lukes J. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei . J. Biol. Chem. 2005;280:2429–2438. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- Xia B., Ke H., Inouye M. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli . Mol. Microbiol. 2001;40:179–188. doi: 10.1046/j.1365-2958.2001.02372.x. [DOI] [PubMed] [Google Scholar]
- Yang Q., Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]