Figure 4. The TBRS/LMS gene ANGPTL4 is a Smad-dependent TGFβ target.
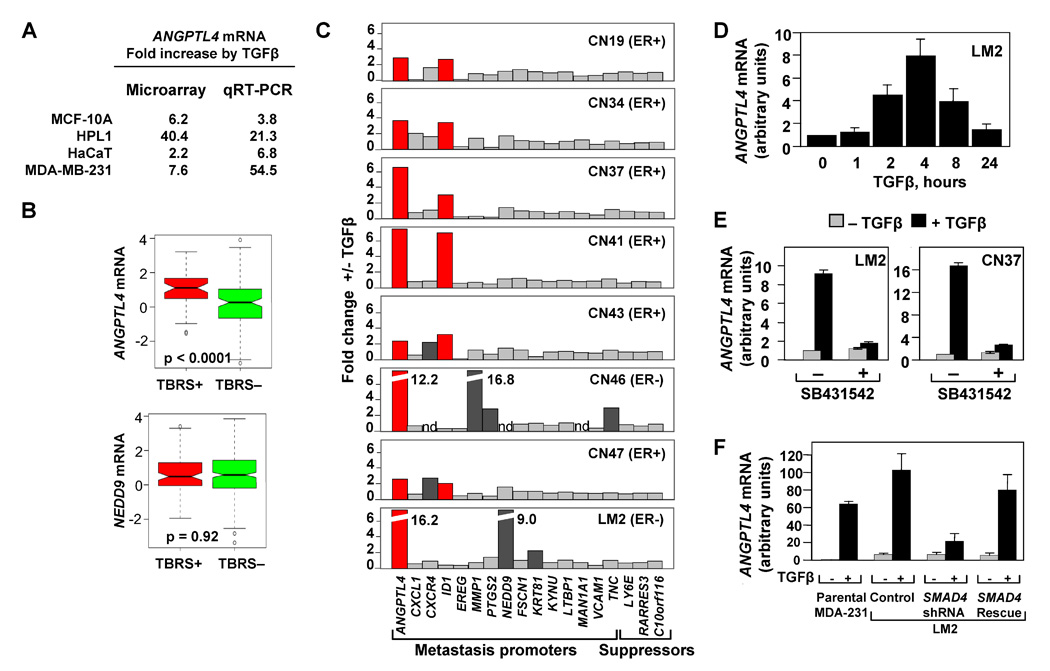
(A) Microarray and qRT-PCR analysis for the four epithelial cell lines treated with TGFβ. Fold change values for the TGFβ induction of ANGPTL4 are indicated.
(B) Box-and-whisker plot comparing ANGPTL4 and NEDD9 TBRS-negative and -positive ER-negative tumors from the MSK/EMC cohorts. P value was calculated using the Wilcoxon rank sum test.
(C) TGFβ-induced changes in the mRNA expression of LMS genes in a panel of clinically derived pleural effusion samples and LM2 cells. Cells were treated with 100 pM of TGFβ for 3 h and analyzed by qRT-PCR using primers for the indicated genes. ER status for each breast cancer patient is designated.
(D) LM2 breast cancer cells were treated with 100pM of TGFβ for the indicated lengths of time and ANGPTL4 mRNA levels were analyzed using qRT-PCR.
(E) Treatment of LM2 (left panel) and pleural effusion-derived CN37 sample (right panel) with TGFβ and the TGFβ-receptor kinase inhibitor, SB431542. qRT-PCR expression levels are shown relative to the untreated control sample.
(F) MDA-231, LM2 control, LM2-Smad4-depleted, and LM2-Smad4-Rescue cell lines were treated with 100 pM TGFβ for 3 h. TGFβ-induced fold changes of ANGPTL4 were analyzed by qRT-PCR analysis, n=3; error bars indicate standard deviation.
