Figure 6. The NotI active site.
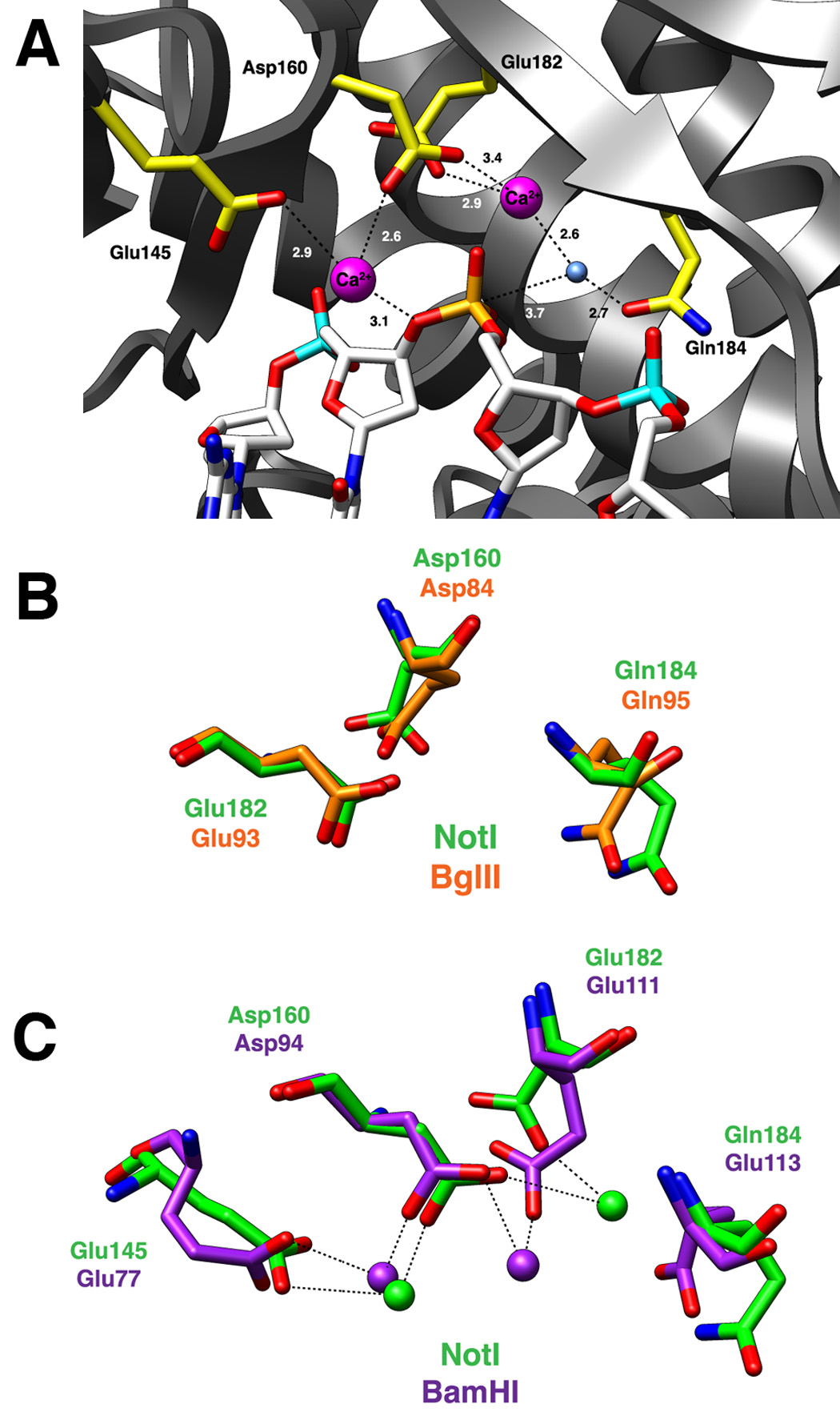
Panel A: Active site residues Glu145, Asp160, Glu182, and Gln184 are colored yellow. The bound DNA is colored white with the scissile phosphate designated orange. The nucleophilic water molecule is shown as a blue sphere and two calcium ions are depicted as magenta spheres. Dashed lines represent interactions between atoms in the active site and numbers indicate distances in angstroms (as average values calculated between the two active sites of the NotI homodimer). Panel B: Superposition of active site residues of the PD…(D/E)xK active site motif from NotI (green) and BglII (orange). Both REases have a glutamine in the general base position typically occupied by a lysine residue. Panel C: Superposition of the two-metal ion active sites of NotI (green) and BamHI (purple). Asp160/Asp94, Glu182/Glu111, and Gln184/Glu113 are residues belonging to the canonical PD…(D/E)xK nuclease motif, while the 3rd acidic residue, Glu145/Glu77, helps coordinate a second metal ion in the active site. The position of this 3rd acidic residue is conserved between the NotI and BamHI enzymes.
