Figure 2.
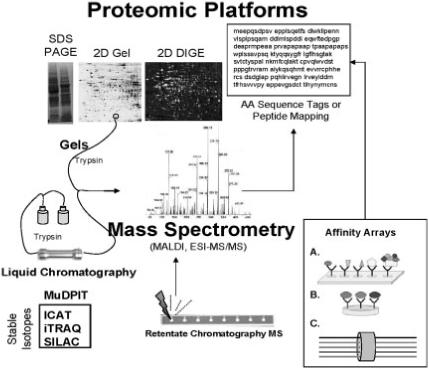
Proteomic platforms for toxicoproteomics studies. Proteomic platforms represent strategies for global separation and identification of proteins. Separations are generally accomplished by gel electrophoresis in toxicoproteomic studies although more recent studies incorporate liquid chromatography-based (LC) platforms such as linear column gradients or multidimensional chromatography (MuDPIT). Use of stable isotopes greatly facilitates protein quantitation (ICAT = isotope coded affinity tags; iTRAQ = isobaric tags for relative and absolute quantitation; SILAC = stable isotope labelling by amino acids in cell culture). Mass spectrometry predominates as a means of protein identification. Identification occurs by peptide mapping or amino acid (AA) sequencing. Retentate chromatography mass spectrometry has been used for rapid profiling of biofluid samples using chemically reactive surfaces for separation and MALDI for generating protein mass spectra (i.e. SELDI technology). Alternatives for MS-based proteomics involve affinity arrays such as (A) antibody arrays, (B) antibody multiplexing and (C) fluorescently tagged antibody bound bead suspensions (i.e. Luminex technology).
