Abstract
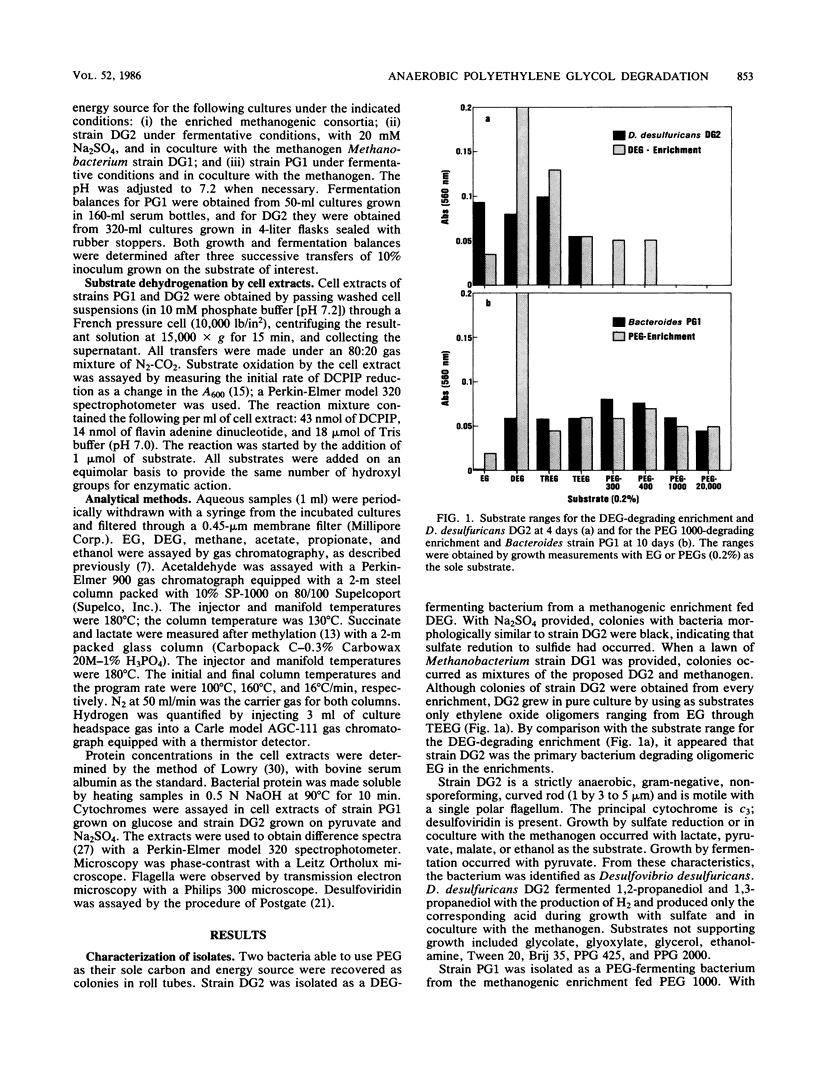
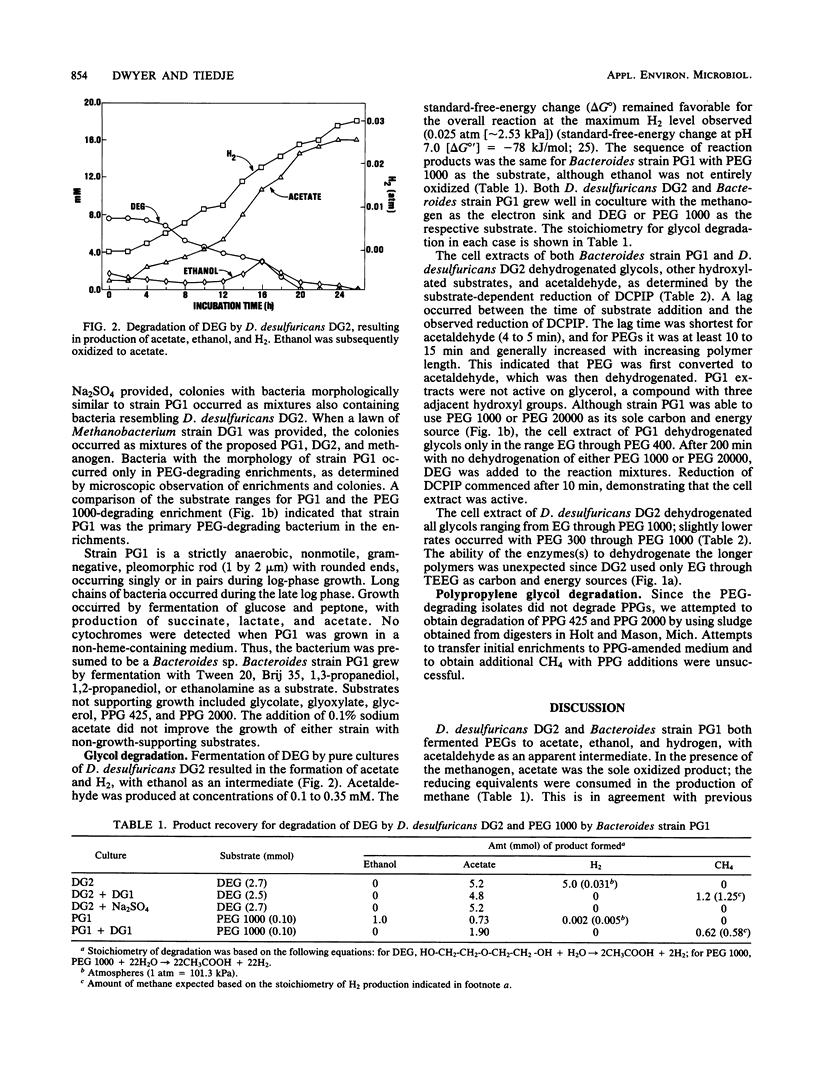
Two anaerobic bacteria were isolated from polyethylene glycol (PEG)-degrading, methanogenic, enrichment cultures obtained from a municipal sludge digester. One isolate, identified as Desulfovibrio desulfuricans (strain DG2), metabolized oligomers ranging from ethylene glycol (EG) to tetraethylene glycol. The other isolate, identified as a Bacteroides sp. (strain PG1), metabolized diethylene glycol and polymers of PEG up to an average molecular mass of 20,000 g/mol [PEG 20000; HO-(CH2-CH2-O-)nH]. Both strains produced acetaldehyde as an intermediate, with acetate, ethanol, and hydrogen as end products. In coculture with a Methanobacterium sp., the end products were acetate and methane. Polypropylene glycol [HO-(CH2-CH2-CH2-O-)nH] was not metabolized by either bacterium, and methanogenic enrichments could not be obtained on this substrate. Cell extracts of both bacteria dehydrogenated EG, PEGs up to PEG 400 in size, acetaldehyde, and other mono- and dihydroxylated compounds. Extracts of Bacteroides strain PG1 could not dehydrogenate long polymers of PEG (greater than or equal to 1,000 g/mol), but the bacterium grew with PEG 1000 or PEG 20000 as a substrate and therefore possesses a mechanism for PEG depolymerization not present in cell extracts. In contrast, extracts of D. desulfuricans DG2 dehydrogenated long polymers of PEG, but whole cells did not grow with these polymers as substrates. This indicated that the bacterium could not convert PEG to a product suitable for uptake.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ABELES R. H., LEE H. A., Jr An intramolecular oxidation-reduction requiring a cobamide coenzyme. J Biol Chem. 1961 Aug;236:2347–2350. [PubMed] [Google Scholar]
- Caskey W. H., Taber W. A. Oxidation of ethylene glycol by a salt-requiring bacterium. Appl Environ Microbiol. 1981 Jul;42(1):180–183. doi: 10.1128/aem.42.1.180-183.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colberg P. J., Young L. Y. Aromatic and Volatile Acid Intermediates Observed during Anaerobic Metabolism of Lignin-Derived Oligomers. Appl Environ Microbiol. 1985 Feb;49(2):350–358. doi: 10.1128/aem.49.2.350-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. P. The biodegradation of polyethylene glycols. Adv Appl Microbiol. 1978;23:173–194. doi: 10.1016/s0065-2164(08)70068-6. [DOI] [PubMed] [Google Scholar]
- Dwyer D. F., Tiedje J. M. Degradation of ethylene glycol and polyethylene glycols by methanogenic consortia. Appl Environ Microbiol. 1983 Jul;46(1):185–190. doi: 10.1128/aem.46.1.185-190.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GASTON L. W., STADTMAN E. R. Fermentation of ethylene glycol by Clostridium glycolicum, sp. n. J Bacteriol. 1963 Feb;85:356–362. doi: 10.1128/jb.85.2.356-362.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C. F., Taber W. A., Zeitoun M. A. Biodegradation of ethylene glycol by a salt-requiring bacterium. Appl Microbiol. 1972 Dec;24(6):911–919. doi: 10.1128/am.24.6.911-919.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines J. R., Alexander M. Microbial degradation of polyethylene glycols. Appl Microbiol. 1975 May;29(5):621–625. doi: 10.1128/am.29.5.621-625.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins L. D., Cook K. A., Cain R. B. Microbial degradation of polyethylene glycols. J Appl Bacteriol. 1979 Aug;47(1):75–85. doi: 10.1111/j.1365-2672.1979.tb01171.x. [DOI] [PubMed] [Google Scholar]
- Nikaido H., Rosenberg E. Y. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J Bacteriol. 1983 Jan;153(1):241–252. doi: 10.1128/jb.153.1.241-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer R., Gerhardt P. Molecular sieving by the Bacillus megaterium cell wall and protoplast. J Bacteriol. 1971 Sep;107(3):718–735. doi: 10.1128/jb.107.3.718-735.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink B., Stieb M. Fermentative degradation of polyethylene glycol by a strictly anaerobic, gram-negative, nonsporeforming bacterium, Pelobacter venetianus sp. nov. Appl Environ Microbiol. 1983 Jun;45(6):1905–1913. doi: 10.1128/aem.45.6.1905-1913.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton D. R., Tiedje J. M. General method for determining anaerobic biodegradation potential. Appl Environ Microbiol. 1984 Apr;47(4):850–857. doi: 10.1128/aem.47.4.850-857.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer R. K., Jungermann K., Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977 Mar;41(1):100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITE D. C. Cytochrome and catalase patterns during growth of Haemophilus parainfluenzae. J Bacteriol. 1962 Apr;83:851–859. doi: 10.1128/jb.83.4.851-859.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts A. Bacterial metabolism of ethylene glycol. Biochim Biophys Acta. 1981 Oct 12;677(2):194–199. doi: 10.1016/0304-4165(81)90085-4. [DOI] [PubMed] [Google Scholar]
- Zalman L. S., Nikaido H. Dimeric porin from Paracoccus denitrificans. J Bacteriol. 1985 Apr;162(1):430–433. doi: 10.1128/jb.162.1.430-433.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]


