Abstract
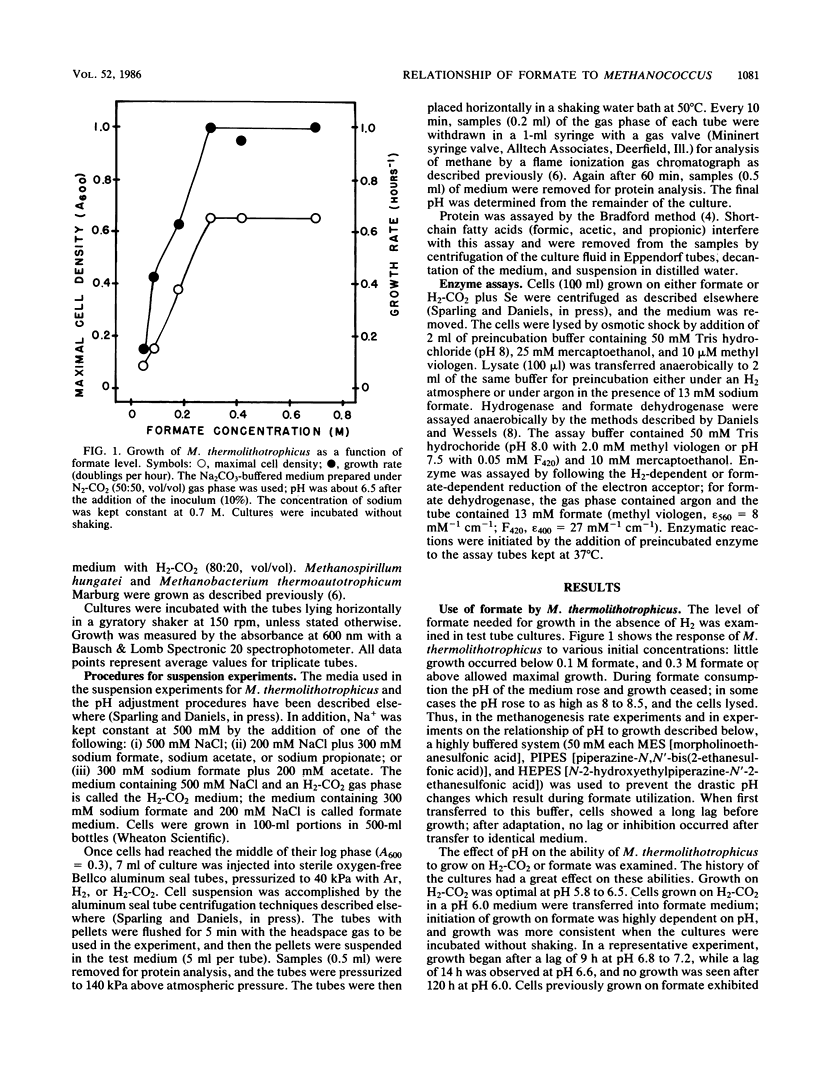
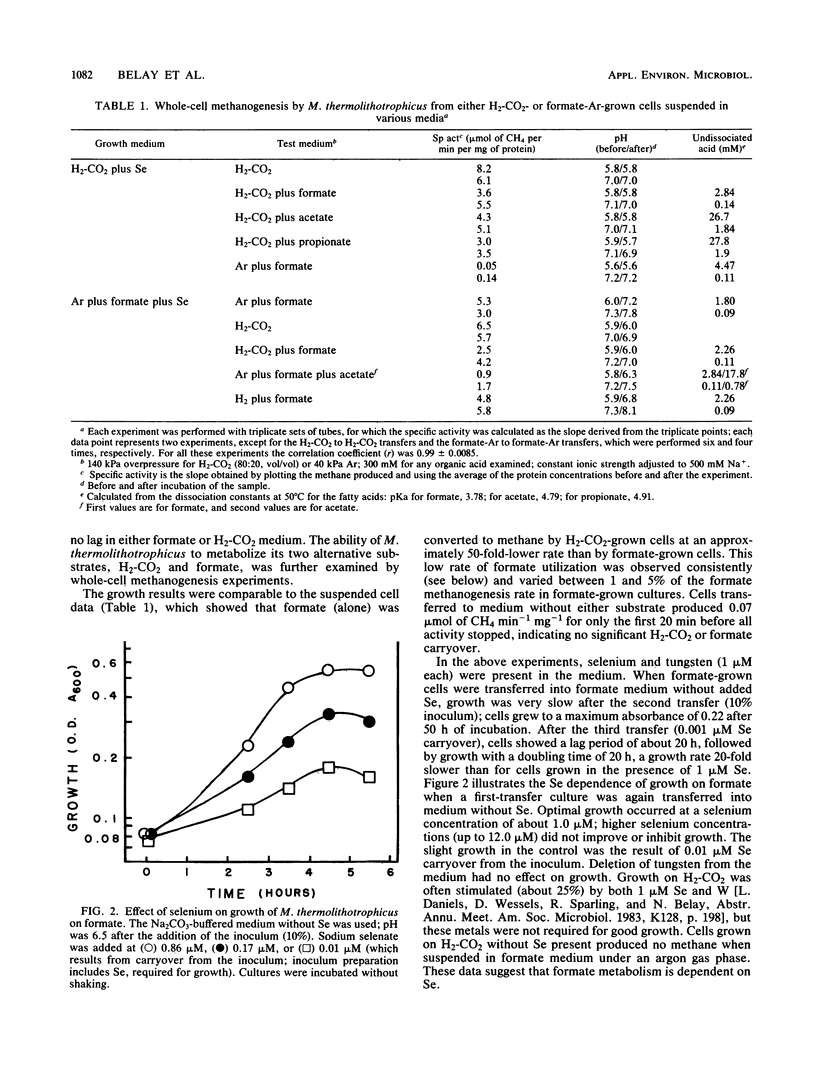
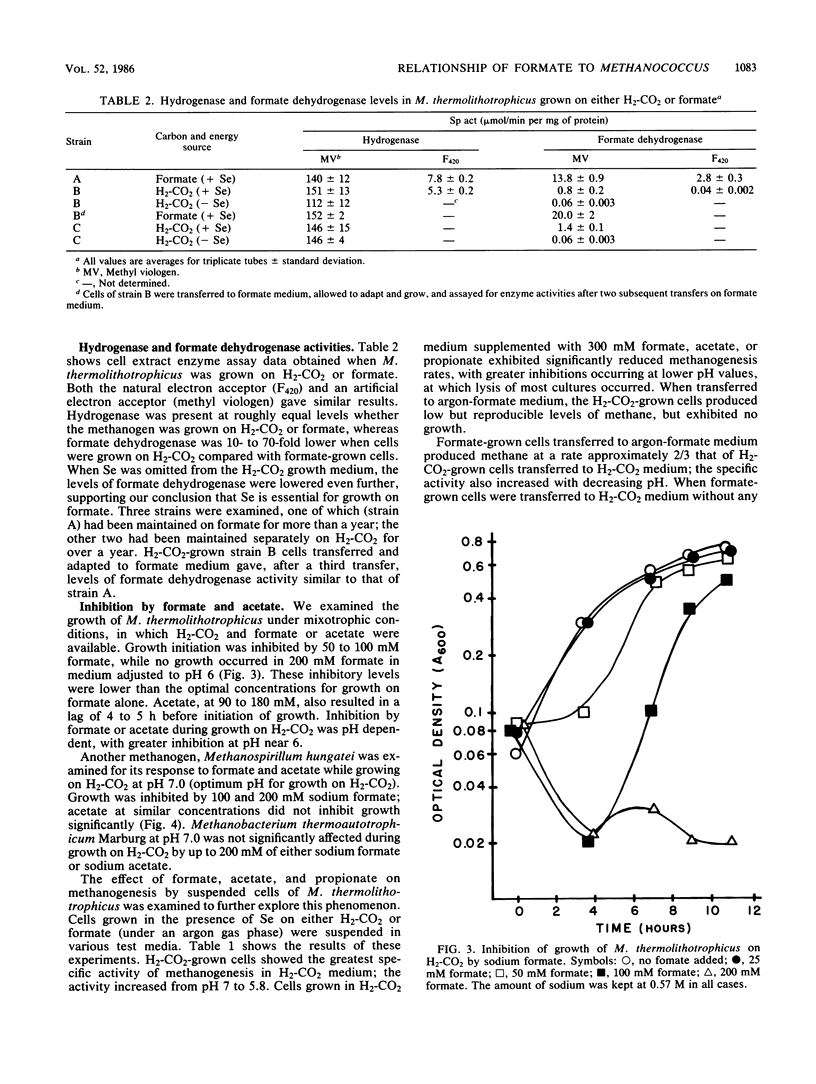
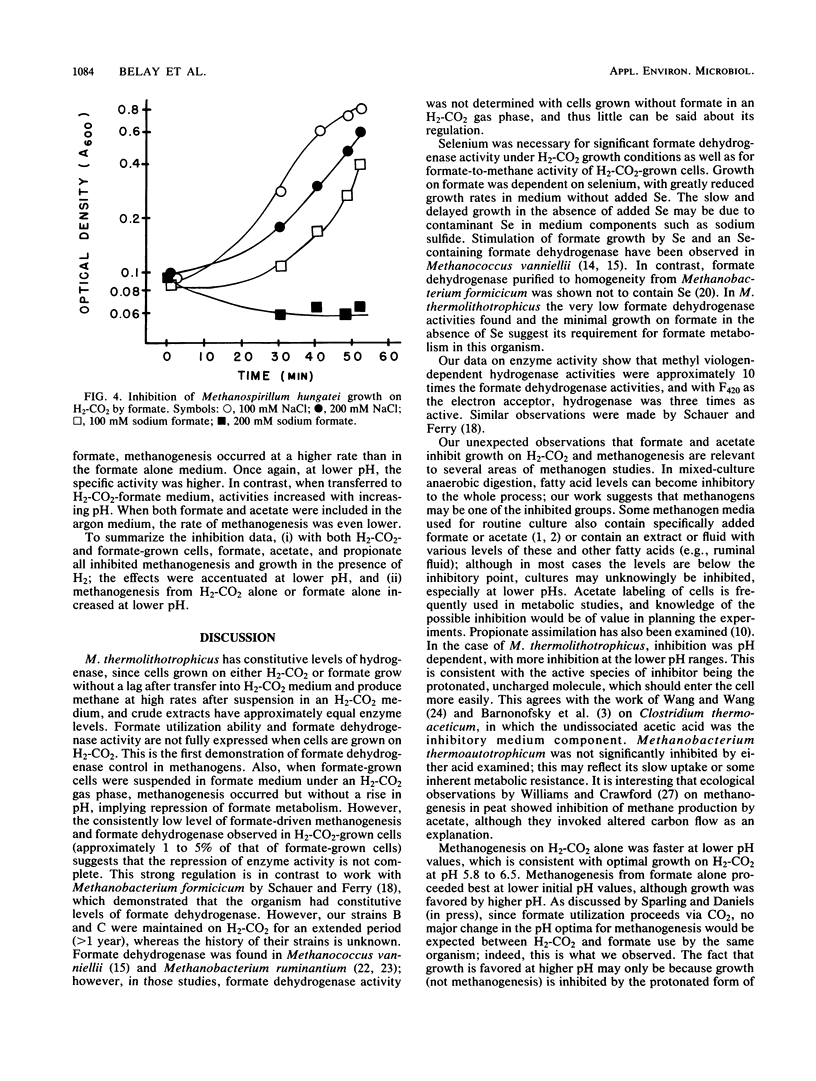
Methanococcus thermolithotrophicus is a methanogenic archaebacterium that can use either H2 or formate as its source of electrons for reduction of CO2 to methane. Growth and suspended-whole-cell experiments show that H2 plus CO2 methanogenesis was constitutive, while formate methanogenesis required adaptation time; selenium was necessary for formate utilization. Cells grown on formate had 20 to 100 times higher methanogenesis rates on formate than cells grown on H2-CO2 and transferred into formate medium. Enzyme assays with crude extracts and with F420 or methyl viologen as the electron acceptor revealed that hydrogenase was constitutive, while formate dehydrogenase was regulated. Cells grown on formate had 10 to 70 times higher formate dehydrogenase activity than cells grown on H2-CO2 with Se present in the medium; when no Se was added to H2-CO2 cultures, even lower activities were observed. Adaptation to and growth on formate were pH dependent, with an optimal pH for both about one pH unit above that optimal for H2-CO2 (pH 5.8 to 6.5). When cells were grown on H2-CO2 in the presence of formate, formate (greater than or equal to 50 mM) inhibited both growth and methanogenesis at pH 5.8 to 6.2, but not at pH greater than 6.6. Both acetate and propionate produced similar inhibition. Formate inhibition was also observed in Methanospirillum hungatei.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Wolfe R. S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl Environ Microbiol. 1976 Dec;32(6):781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baronofsky J. J., Schreurs W. J., Kashket E. R. Uncoupling by Acetic Acid Limits Growth of and Acetogenesis by Clostridium thermoaceticum. Appl Environ Microbiol. 1984 Dec;48(6):1134–1139. doi: 10.1128/aem.48.6.1134-1139.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Daniels L., Belay N., Rajagopal B. S. Assimilatory reduction of sulfate and sulfite by methanogenic bacteria. Appl Environ Microbiol. 1986 Apr;51(4):703–709. doi: 10.1128/aem.51.4.703-709.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels L., Sparling R., Sprott G. D. The bioenergetics of methanogenesis. Biochim Biophys Acta. 1984 Sep 6;768(2):113–163. doi: 10.1016/0304-4173(84)90002-8. [DOI] [PubMed] [Google Scholar]
- Daniels L., Wessels D. A method for the spectrophotometric assay of anaerobic enzymes. Anal Biochem. 1984 Aug 15;141(1):232–237. doi: 10.1016/0003-2697(84)90450-0. [DOI] [PubMed] [Google Scholar]
- Daniels L., Zeikus J. G. One-carbon metabolism in methanogenic bacteria: analysis of short-term fixation products of 14CO2 and 14CH3OH incorporated into whole cells. J Bacteriol. 1978 Oct;136(1):75–84. doi: 10.1128/jb.136.1.75-84.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G., Stupperich E., Thauer R. K. Acetate assimilation and the synthesis of alanine, aspartate and glutamate in Methanobacterium thermoautotrophicum. Arch Microbiol. 1978 Apr 27;117(1):61–66. doi: 10.1007/BF00689352. [DOI] [PubMed] [Google Scholar]
- Jones J. B., Dilworth G. L., Stadtman T. C. Occurrence of selenocysteine in the selenium-dependent formate dehydrogenase of Methanococcus vannielii. Arch Biochem Biophys. 1979 Jul;195(2):255–260. doi: 10.1016/0003-9861(79)90351-5. [DOI] [PubMed] [Google Scholar]
- Jones J. B., Stadtman T. C. Methanococcus vannielii: culture and effects of selenium and tungsten on growth. J Bacteriol. 1977 Jun;130(3):1404–1406. doi: 10.1128/jb.130.3.1404-1406.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G. B., Roth L. A., van den Berg L., Clark D. S. Characterization of a strain of Methanospirillum hungatti. Can J Microbiol. 1976 Sep;22(9):1404–1410. doi: 10.1139/m76-208. [DOI] [PubMed] [Google Scholar]
- Schauer N. L., Brown D. P., Ferry J. G. Kinetics of Formate Metabolism in Methanobacterium formicicum and Methanospirillum hungatei. Appl Environ Microbiol. 1982 Sep;44(3):549–554. doi: 10.1128/aem.44.3.549-554.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Composition of the coenzyme F420-dependent formate dehydrogenase from Methanobacterium formicicum. J Bacteriol. 1986 Feb;165(2):405–411. doi: 10.1128/jb.165.2.405-411.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. FAD requirement for the reduction of coenzyme F420 by formate dehydrogenase from Methanobacterium formicicum. J Bacteriol. 1983 Aug;155(2):467–472. doi: 10.1128/jb.155.2.467-472.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Metabolism of formate in Methanobacterium formicicum. J Bacteriol. 1980 Jun;142(3):800–807. doi: 10.1128/jb.142.3.800-807.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprott G. D., Jarrell K. F. K+, Na+, and Mg2+ content and permeability of Methanospirillum hungatei and Methanobacterium thermoautotrophicum. Can J Microbiol. 1981 Apr;27(4):444–451. doi: 10.1139/m81-067. [DOI] [PubMed] [Google Scholar]
- Tzeng S. F., Wolfe R. S., Bryant M. P. Factor 420-dependent pyridine nucleotide-linked hydrogenase system of Methanobacterium ruminantium. J Bacteriol. 1975 Jan;121(1):184–191. doi: 10.1128/jb.121.1.184-191.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzing S. F., Bryant M. P., Wolfe R. S. Factor 420-dependent pyridine nucleotide-linked formate metabolism of Methanobacterium ruminantium. J Bacteriol. 1975 Jan;121(1):192–196. doi: 10.1128/jb.121.1.192-196.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOLIN E. A., WOLIN M. J., WOLFE R. S. FORMATION OF METHANE BY BACTERIAL EXTRACTS. J Biol Chem. 1963 Aug;238:2882–2886. [PubMed] [Google Scholar]
- Wang G., Wang D. I. Elucidation of Growth Inhibition and Acetic Acid Production by Clostridium thermoaceticum. Appl Environ Microbiol. 1984 Feb;47(2):294–298. doi: 10.1128/aem.47.2.294-298.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer P. J., Zeikus J. G. Acetate assimilation pathway of Methanosarcina barkeri. J Bacteriol. 1979 Jan;137(1):332–339. doi: 10.1128/jb.137.1.332-339.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer P. J., Zeikus J. G. Acetate metabolism in Methanosarcina barkeri. Arch Microbiol. 1978 Nov 13;119(2):175–182. doi: 10.1007/BF00964270. [DOI] [PubMed] [Google Scholar]
- Williams R. T., Crawford R. L. Methane production in Minnesota peatlands. Appl Environ Microbiol. 1984 Jun;47(6):1266–1271. doi: 10.1128/aem.47.6.1266-1271.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]


