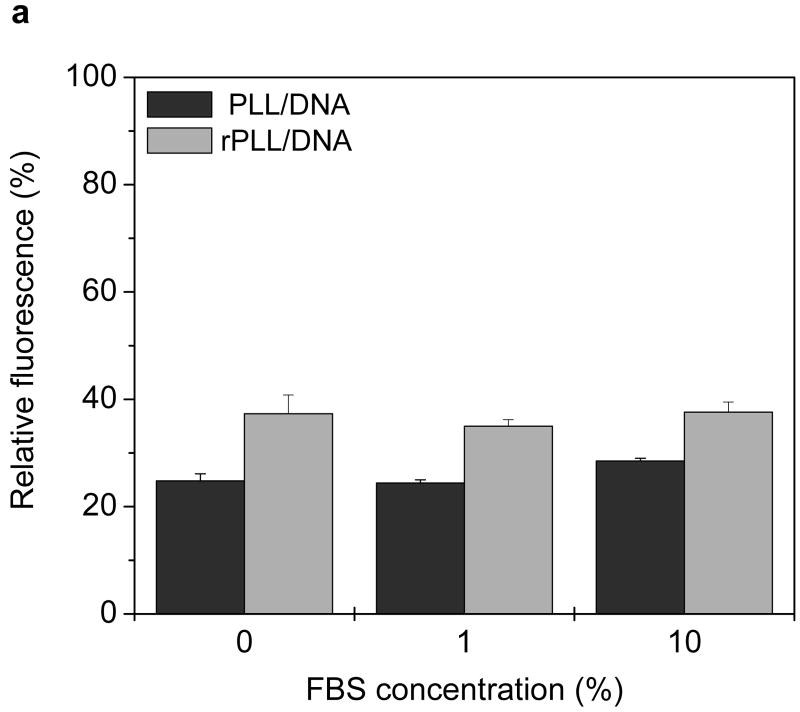
Figure 4. Effect of serum on the stability of pDNA complexes.
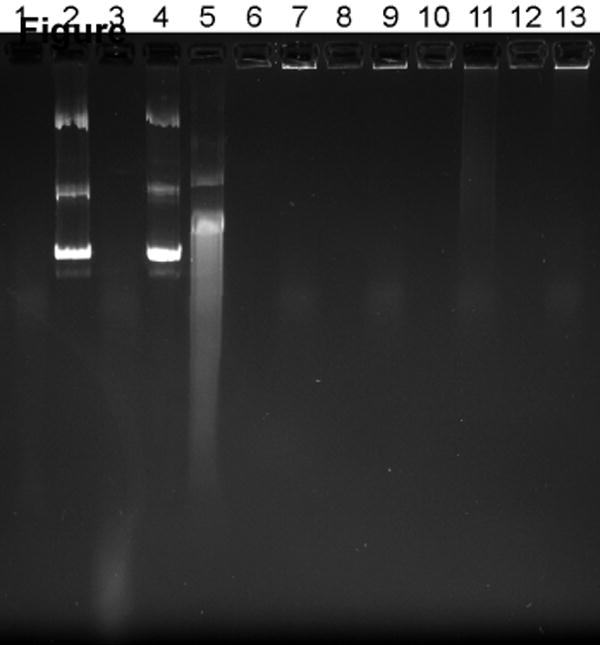
(a) Fluorescence recovery assay. Twenty μL of complexes containing YOYO-1 labeled pDNA were incubated with 80 μL of RPMI containing 0, 1, and 10% FBS for 4 h at 37°C. Fluorescence of YOYO-1 labeled pDNA was measured and normalized to fluorescence intensity of free pDNA. Results are expressed as mean ± SD of triplicate samples. (b) Agarose gel electrophoresis assay. Complexes were prepared using non-labeled pDNA and subjected to treatment as in 4a. Samples containing 0.2 μg pDNA were run on a 0.8% agarose gel and pDNA was stained with ethidium bromide and visualized under UV illumination. Lane 1, RPMI + 10% FBS; lane 2, pDNA + deionized water (DW); lane 3, pDNA + DW + 10% FBS; lane 4, pDNA + RPMI; lane 5, pDNA + RPMI + 10% FBS; lane 6, PLL/DNA + DW; lane 7, PLL/DNA + DW + 10% FBS; lane 8, PLL/DNA + RPMI; lane 9, PLL/DNA + RPMI + 10% FBS; lanes 10-13, rPLL/DNA with respective treatments indicated in lanes 6-9.