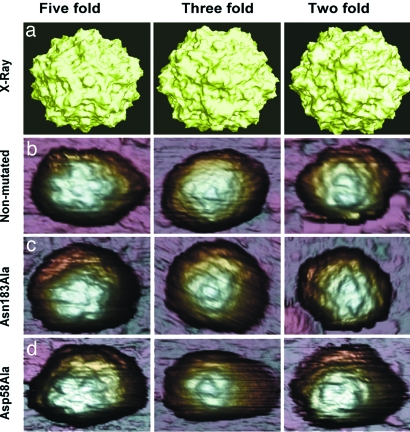
Fig. 2.
MVM virions imaged by AFM and viewed along an S5 (Left), S3 (Center), or S2 (Right) symmetry axis. (a) Molecular surface model of the nonmutated MVM virion derived from crystallographic data. (b–d) AFM images corresponding to nonmutated virion (b), Asn183Ala mutant (c), and Asp58Ala mutant (d). The MVM topography appears laterally expanded because of the usual tip-sample dilation effects. Only those particles with a symmetry axis at, or very close to, the top (center of the image) were selected for indentation. Most particles showed a symmetry axis clearly off-center, or no recognizable symmetry, and were discarded.