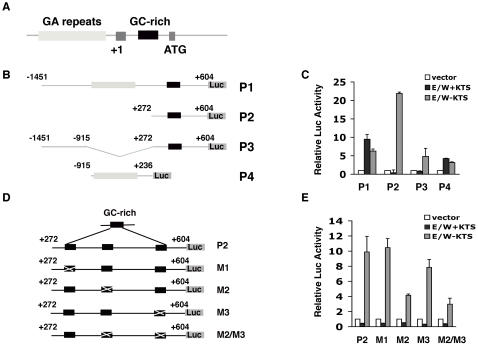
Figure 2. Identification of EWS-WT1(+KTS) and (−KTS) responsive elements in the human ENT4 promoter.
A. Schematic representation of ENT4 gene. The dark grey box indicated by +1 represents the first non-coding exon and the transcription start site (+1). The second dark grey box marked by ATG represents exon 2 containing the starting ATG. The light grey box represents the GA-repeat sequence. The black box represents the GC-rich sequence. B. Schematics showing the luciferase reporter constructs (P1–P4) containing different fragments of the ENT4 promoter. P3 promoter construct contains the deletion of GA-repeat sequences (−915 to +272). The numbers are relative to the transcription start (+1). C. Direct transcriptional activation of ENT4 promoters by EWS/WT1. U2OS cells were co-transfected with either pcDNA3-EWS/WT1 (− or +KTS; 0.5 µg) or with pcDNA3 along with the promoter-reporter constructs (P1, P2, P3 or P4; 0.5 µg) and Renilla luciferase (0.1 µg) using FuGENE 6. Luciferase activity was measured at 48 hrs post-transfection and expressed as relative luciferase activity compared to the empty vector. Data represent the mean±S.D. from three independent experiments. D. Schematics showing the three putative EWS/WT1(−KTS) binding sites (black boxes) within the GC-rich region of the P2 promoter and the mutated derivatives, M1, M2, M3 and M2/M3 constructs (see Materials and Methods for detail). E. Identification of the EWS/WT1(−KTS) responsive element M2. Luciferase-reporter assay was performed as in (C) except with the promoter-reporter constructs shown in (D). Data represent the mean±S.D. from three independent experiments.