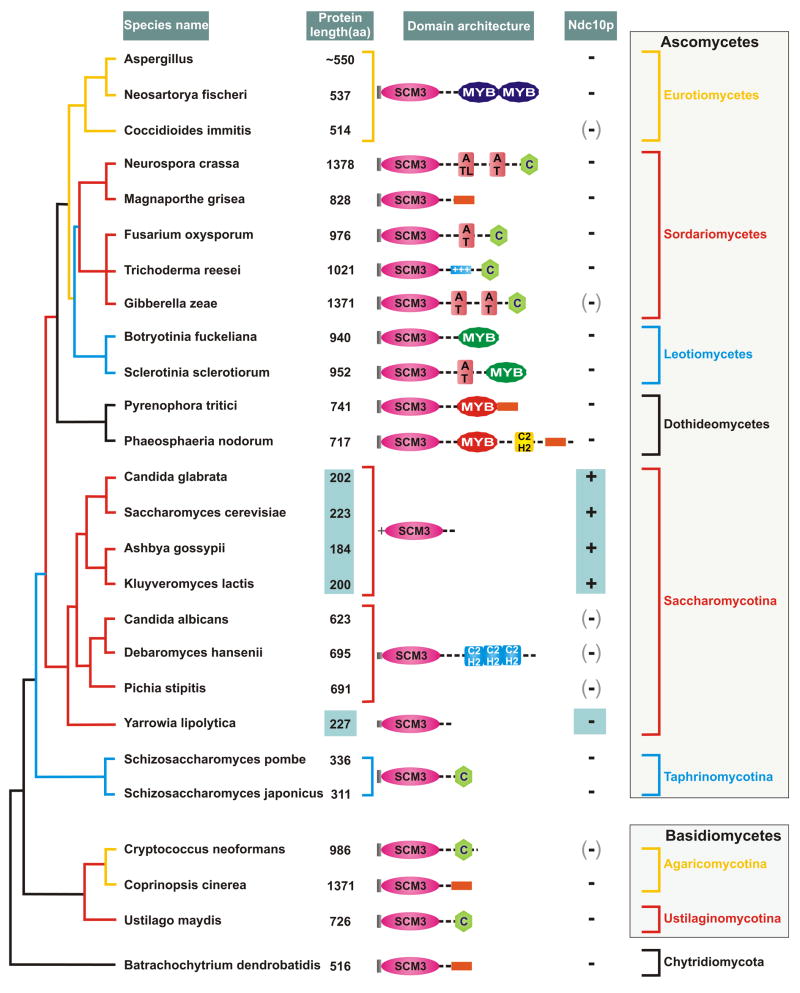
Figure 1. Diversity of domain architectures of Scm3p across fungal phylogeny.
The domain architectures of Scm3p proteins have been superimposed on a maximum-likelihood phylogenetic tree of fungi based on six conserved proteins. The major lineages shown in the figure are supported by 80% or greater relative logarithm likelihood bootstrap support. The proteins are not rendered to scale because of their enormous size difference, though the conserved domains are approximately rendered to size. The actual size of each protein is shown to its left and the small versions are boxed. The notations are: C- Cysteine cluster; the orange box represents positively charged segments. Notably, the N-termini of Scm3p orthologs across much of the fungal tree begin with a conserved block with the signature MxxP (shown as a grey block), but those of most members belonging to Saccharomycotina lack this signature. Instead they possess a distinctive positively charged helical segment indicated with a “+”. There is some uncertainty regarding the predicted gene structure, and hence the predicted protein of Cryptococcus neoformans. The different independently acquired versions of the MYB domain are shaded differently. The Ndc10 column reflects the presence or absence of orthologs of the Ndc10p protein, where +: presence, −: absence and (-): species with a homologous GCR1 domain, either in paralogous transcription factors or crypton family transposons, but no Ndc10 ortholog. All domains noted here were detected with e-values <.01 in profile and HMM searches. They were confirmed through reciprocal searches using PSI-BLAST that recovered known representatives of each of the depicted domains with e <.01 prior to convergence.