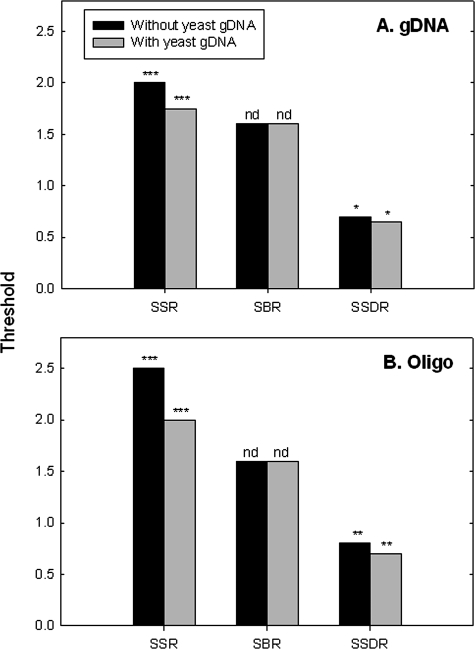
FIG. 5.
Effects of background DNA on the determination of SSR, SBR, and SSDR thresholds. Five hundred nanograms of S. oneidensis MR-1 gDNA (A) and 10 pg for each synthesized oligonucleotide (oligo) (B) were spiked into 1.0 μg of yeast gDNA. For synthesized oligonucleotide targets, the yeast gDNA was first labeled and then mixed with the spiked oligonucleotides. S. oneidensis MR-1 gDNA was first mixed with the yeast gDNA and then labeled together. The significance is shown on the top of each column, with the following notations: nd, no difference; one asterisk, P < 0.10; two asterisks, P < 0.05; and three asterisks, P < 0.01 (the Student t test) when thresholds with background DNA were compared to those without background DNA.