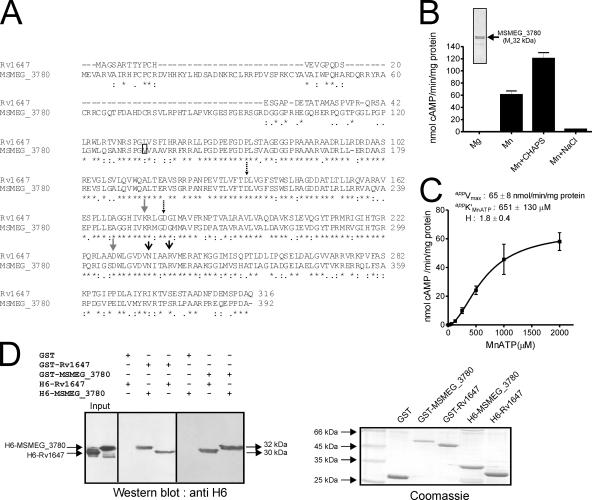
FIG. 2.
Expression, purification, and characterization of MSEMG_3786 adenylyl cyclase and interaction with Rv1647. (A) Sequence similarity of the Rv1647 and the MSMEG_3780 proteins. The boxed residue represents the annotated start of MSMEG_3780 according to the Sanger Centre annotation. The predicted amino acid sequences were aligned using ClustalW. Dashed arrows represent substrate-specifying residues; gray arrows represent metal-binding residues; black arrows represent transition-state stabilizing residues. (B) Purified MSMEG_3780 (∼500 ng) was assayed in the presence of either 10 mM Mg or 10 mM Mn as a free metal along with 1 mM ATP. CHAPS (0.5%) or NaCl (500 mM) was added in some assays, along with 10 mM Mn and 1 mM MnATP as a substrate, and assays were carried out at pH 9. The inset shows purified MSMEG_3780 used for adenylyl cyclase assays. Values represent the means ± SEM of duplicate determinations, with each assay being performed three times. (C) MSMEG_3780 activity was assayed in the presence of various concentrations of MnATP in the presence of 10 mM free Mn. The level of cAMP produced was measured by radioimmunoassay. All data shown represent the means ± SEM of triplicate determinations, with each assay being performed twice. (D) A GST pull-down assay was performed as described in Materials and Methods. Hexahistidine-tagged proteins that interacted with the GST fusion protein were detected by Western blot analysis using an anti-His6 antibody. An equal aliquot of proteins that were taken for interaction was subjected to SDS-polyacrylamide gel electrophoresis, and the gel was stained to ensure that approximately equal amounts of protein were used for the experiment. The result shown is representative of experiments performed twice.