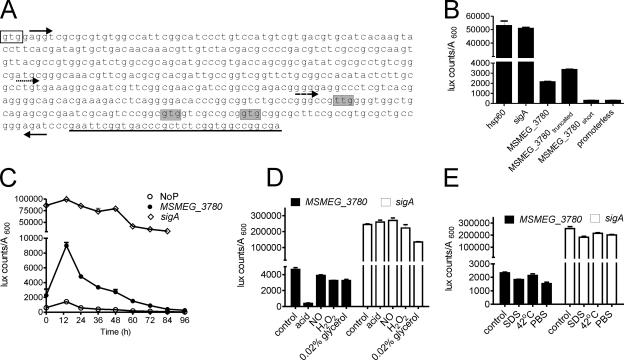
FIG. 5.
Identification of the MSMEG_3780 promoter and its activity during growth and stress conditions. (A) Sequence of a part of the MSMEG_3780 gene indicating promoters used for analysis. Forward arrows indicate the 5′ end of the MS_3780, MS_3780truncated (dotted arrow), or MS_3780short (dashed arrow) promoter, and the reverse arrow indicates the 3′ ends of the promoter constructs. The clear box indicates the translational start codon as annotated by TIGR, while shaded nucleotides indicate alternative start codons based on the promoter analysis. Underlined bases indicate sequences that encode the beginning of the catalytic domain of MSMEG_3780. (B) Regions containing the putative promoter of MSMEG_3780 were cloned upstream of the luxAB gene, and luciferase activity was measured. The activities of the hsp60 and sigA promoters and a promoterless construct were also monitored. (C) Activities of the promoterless construct (NoP) and MSMEG_3780 and sigA promoters during growth. M. smegmatis cells were transformed with the indicated plasmids, and luciferase activity was monitored as described in Materials and Methods. (D) M. smegmatis cells were cultured for 30 h, and cells were resuspended in medium as indicated and cultured for another 5 h, following which luciferase activity was monitored. (E) M. smegmatis cells were cultured for 30 h, and cells were resuspended in medium for another 1.5 h at 37°C or at 42°C. Alternatively, cells were resuspended in medium containing 0.05% SDS or in PBS and cultured for 1.5 h, followed by the estimation of luciferase activity. All data shown are the means ± SEM of duplicate determinations of experiments performed three times.