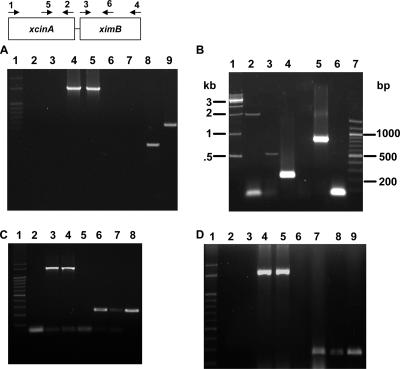
FIG. 3.
Analysis of xcinA and ximB mRNA by RT-PCR. (A) Comparison of X. nematophila strain 19060 and xenocin-sensitive isolate X(sensitive) after induction with mitomycin C. Lane 1, markers; lanes 2 and 3, RNA from X(sensitive) and strain 19060, respectively, without reverse transcriptase; lanes 4 and 5, control 16S RNA from X(sensitive) and strain 19060, respectively; lanes 6 and 7, RNA from X(sensitive) amplified with primers 5 and 2 (lane 6) and with primers 5 and 6 (lane 7); lanes 8 and 9, RNA from X. nematophila 19060 amplified with primers 5 and 2 (lane 8) and with primers 5 and 6 (lane 9). (B) X. nematophila cells were induced with mitomycin C for 3 h, and total RNA was isolated. Lane 1, 1-kb ladder; lane 2, primers 1 and 2; lane 3, primers 5 and 6; lane 4, primers 5 and 2; lane 5, primers 3 and 4; lane 6, primers 1 and 4; lane 7, 100-bp ladder. (C) Analysis of xcinA mRNA induced by 25 mM Desferal. Lane 1, 100-bp ladder; lane 2, RNA from induced cells without reverse transcriptase, amplified with primers 5 and 2; lanes 3 and 4, 16S RNA from uninduced and induced loading controls, respectively; lanes 5 and 6, RNA from uninduced (lane 5) and induced (lane 6) cells amplified with primers 5 and 2; lanes 7 and 8, RNA from uninduced (lane 7) and induced (lane 8) cells amplified with primers 3 and 6. (D) Analysis of xcinA mRNA by heat induction. Lane 1, 100-bp ladder; lanes 2 and 3, RNA from uninduced and induced cells without reverse transcriptase, respectively; lanes 4 and 5, 16S RNA controls; lanes 6 and 7, RNA from cells grown at 30°C (lane 6) and at 37°C (lane 7) amplified with primers 5 and 2; lanes 8 and 9, RNA from cells grown at 30°C (lane 8) and at 37°C (lane 9) amplified with primers 3 and 6.