Abstract
We report here that a cancer gene therapy protocol using a combination of IL-12, pro-IL-18, and IL-1β converting enzyme (ICE) cDNA expression vectors simultaneously delivered via gene gun can significantly augment antitumor effects, evidently by generating increased levels of bioactive IL-18 and consequently IFN-γ. First, we compared the levels of IFN-γ secreted by mouse splenocytes stimulated with tumor cells transfected with various test genes, including IL-12 alone; pro-IL-18 alone; pro-IL-18 and ICE; IL-12 and pro-IL-18; and IL-12, pro-IL-18, and ICE. Among these treatments, the combination of IL-12, pro-IL-18, and ICE cDNA resulted in the highest level of IFN-γ production from splenocytes in vitro, and similar results were obtained when these same treatments were delivered to the skin of a mouse by gene gun and IFN-γ levels were measured at the skin transfection site in vivo. Furthermore, the triple gene combinatorial gene therapy protocol was the most effective among all tested groups at suppressing the growth of TS/A (murine mammary adenocarcinoma) tumors previously implanted intradermally at the skin site receiving DNA transfer by gene gun on days 6, 8, 10, and 12 after tumor implantation. Fifty percent of mice treated with the combined three-gene protocol underwent complete tumor regression. In vivo depletion experiments showed that this antitumor effect was CD8+ T cell-mediated and partially IFN-γ-dependent. These results suggest that a combinatorial gene therapy protocol using a mixture of IL-12, pro-IL-18, and ICE cDNAs can confer potent antitumor activities against established TS/A tumors via cytotoxic CD8+ T cells and IFN-γ-dependent pathways.
Recent advances in our understanding of cytokine biology and tumor immunology have made it possible to evaluate new cytokine based anticancer therapies (1–3). Among many cytokines evaluated as anticancer agents, interleukin-12 (IL-12) confers potent antitumor activity in murine tumor models (4–8). IL-12 is a heterodimeric protein consisting of two subunits (p35 and p40) that facilitates the Th1-type immune responses (9, 10). The antitumor effects of IL-12 are mediated by activation of cytotoxic T lymphocytes (CTL) as well as NK cells (11–13), and by the induction of IFN-γ production by T cells and NK cells (6, 14). IFN-γ alone, however, is not sufficient to induce tumor regression, and antibodies to IFN-γ do not completely block the antitumor effect of IL-12 (6, 14). Nevertheless, IFN-γ is involved in a spectrum of the antitumor effects of IL-12 (14). For instance, IFN-γ induced by IL-12 was shown to up-regulate MHC class I and II expression on tumor cells, to activate NK cells and macrophages, to help generate CD8+ cytotoxic T cells, and to induce the production of interferon-inducible protein 10 (IP-10) and monokine induced by IFN-γ (7, 15, 16).
Interleukin-18 (IL-18), originally termed IFN-γ-inducing factor, is also a costimulatory factor in the activation of Th1 cells. IL-18 induces IFN-γ production in both T cells and NK cells (17, 18, 19). In addition, IL-18 induces T cells to produce granulocyte-macrophage colony-stimulating factor, augments IL-2 production by antigen- or anti-CD3-stimulated murine Th1 clones, induces IL-2-dependent proliferation of these stimulated cells, and enhances the cytolytic activity of T cells and NK cells (19–24). IL-18 is synthesized as a biologically inactive precursor molecule (pro-IL-18). To generate the active form of IL-18, pro-IL-18 needs to be cleaved by the intracellular cysteine protease, IL-1β converting enzyme (ICE), at the Asp-X processing site (25, 26). Cleavage of pro-IL-18 by ICE facilitates secretion of the active form of IL-18 (26). IL-18 can inhibit tumor growth in some murine tumor systems, but regression of established tumor by IL-18 gene therapy alone has not been demonstrated (27–31). Moreover, in these studies, only the pro-IL-18 gene was administered, and the ICE gene was not taken into consideration. Thus, in the present study, dual transfection of pro-IL-18 and ICE cDNA was performed from the more physiological viewpoint, instead of transfection of genetically engineered mature form of IL-18 cDNA.
Based on the findings that both IL-12 and IL-18 can augment cytotoxic activities of CTL and NK cells, and can synergize in the production of IFN-γ (21, 32), a combination of IL-12 and IL-18 was demonstrated to confer a superior antitumor activity over either IL-12 or IL-18 treatment alone (30, 31). However, the systemic administration of recombinant IL-12 and IL-18 proteins, though effective in tumor growth inhibition, resulted in death of all animals because of toxicity (30). Systemic administration of IL-12 also was associated with severe dose-dependent toxicity in patients during the first human trial (33). The localized transfer of cytokine genes may circumvent some of the toxicity of systemic IL-12 delivery and provide adequate local cytokine levels for immune cell activation (5, 6, 8). It also has been documented that a number of individual cytokine genes are effective in gene therapy studies in specific mouse tumor systems. However, it is not clear whether a combinatorial approach of using multiple cytokine genes may offer improved efficiency in antitumor activity.
Particle-mediated gene transfer was used to deliver cytokine genes to the tumor microenvironment. It has been shown that gene gun-mediated transfection of tumor cells ex vivo or the skin overlying tumor in vivo with several cytokine genes resulted in tumor regression (8, 34, 35). This technique allows the delivery of multiple genes. In this study, we evaluated and characterized the combination of IL-12, pro-IL-18 and ICE cDNA expression vectors for cancer gene therapy using particle-mediated gene transfer.
Materials and Methods
Mice and Cell Lines.
BALB/c female mice between 6 and 8 weeks of age were purchased from Harlan–Sprague–Dawley and were housed in an American Association for the Accreditation of Laboratory Animal Care-accredited facility under isothermal conditions and allowed access to food and water ad libitum. Five cell lines were used in this study: TS/A (murine mammary adenocarcinoma), Lewis lung carcinoma (LLC), B16 (murine melanoma), RAW 264.7 (murine macrophage), and COS-7 cells. TS/A is a moderately immunogenic tumor and is MHC class I positive (H-2Dd, H-2Kd). This cell line was originally obtained from G. Forni (University of Turin, Milan). The cell cultures were maintained in RPMI medium 1640 (BioWhittaker) supplemented with 10% heat-inactivated fetal bovine serum (Sigma), 2 mM l-glutamine, 1 mM sodium pyruvate, 1% minimal Eagle’s medium nonessential amino acid, 100 unit/ml penicillin, and 100 μg/ml streptomycin (BioWhittaker) in a humidified atmosphere of 5% CO2 at 37°C.
cDNA Expression Plasmid Construction.
The murine IL-12 expression plasmid, pNGVL3-mIL-12, was constructed in a backbone plasmid containing a cytomegalovirus immediate/early enhancer promoter, intron A, and a kanamycin selection gene. The p35 and p40 subunits, separated by an internal ribosomal entry site, were subcloned into the multiple cloning site of pNGVL-3 (National Gene Vector Laboratory, University of Michigan, Ann Arbor, MI). The backbone plasmid, pNGVL-3, was used as a control vector. To construct a plasmid expressing pro-IL-18 or ICE cDNA, the full length of murine pro-IL-18 or ICE cDNA was obtained from total RNA of C57BL/6 mouse spleen cells by reverse transcription–(RT) PCR using a RT-PCR kit (Takara, Tsukuba, Japan). The primers for RT-PCR were designed as follows: pro-IL-18, (sense) 5′-ACCTTCCAAATCACTTCCTC-3′ and (antisense) 5′-CAGGCGAGGTC ATCACAAGG-3′; mICE, (sense) 5′-CTGCGGTGTAGAAAAGAAACG-3′ and (antisense) 5′-GGCACGATTCTCAGCATAGG-3′. The PCR product of pro-IL-18 was subcloned into pNGVL-3 (pNGVL-3-mIL-18), and that of ICE was directly subcloned into the expression vector pCR3.1 (Invirogen, Carlsbad, CA) by TA cloning (pCR3.1-ICE).
DNA-Gold Particle Preparation.
We used a helium-pulse PowderJect VC-1 gene gun device that was designed by D. McCabe (Agracetus, Middleton, WI). Plasmid DNA was precipitated onto 1.5–2.0 μm gold particles as described (8). In this experiment, 28 mg of gold particles and 105 μg of plasmid DNA were used. Each pulse of helium expels the DNA-coated gold beads from a single 0.5-inch segment of gold bead-coated tubing and results in a delivery of 0.5 mg of gold and 1.875 μg of plasmid DNA per transfection.
In Vitro Gene Transfer and IL-18 Bioassay.
For in vitro transfection, 2.5 × 106 COS-7 or TS/A cells were suspended in 30 μl of RPMI medium 1640 and were spread into the target size in a 35-mm dish. Transfections with cytokine cDNA were performed by using a gene gun with a 250-psi helium pulse (1 psi = 6.89 kPa). Cells then were recounted by using the trypan blue dye exclusion assay, and 1 × 106 viable cells were placed in 2.0 ml of culture medium. Culture supernatants were harvested at 24 hr posttransfection for use in the IL-18 bioassay. The relative bioactivity of IL-18 was determined by the ability of cell supernatants to augment IFN-γ production in vitro (18). In brief, mouse splenocytes (2 × 106) were cocultured with Con A (1.25 μg/ml) in 24-well plates for 48 hr. Supernatants obtained from transfected cells were added to cell suspensions of Con A-primed splenocytes (1 × 106) in 96-well plates for 24 hr. The supernatants were collected and assayed by ELISA to detect IFN-γ production.
Murine Tumor Model and in Vivo Treatment Protocol.
Mice were shaved in the abdominal area and were injected intradermally with 1 × 105 tumor cells in 50 μl of PBS. Mouse skin overlying and surrounding the target tumor was transfected in vivo by using a 400-psi blast with cytokine cDNA expression vectors and control plasmid (pNGVL-3) on days 6, 8, 10, and 12 after tumor implantation. There were six groups of gene gun treatments, which included: (i) mIL-12 cDNA alone; (ii) murine pro-IL-18 cDNA alone; (iii) murine pro-IL-18 and ICE cDNA (pro-IL-18/ICE); (iv) mIL-12 and pro-IL-18 cDNA (IL-12/pro-IL-18); (v) mIL-12, pro-IL-18 and ICE cDNA (IL-12/pro-IL-18/ICE); and (vi) control vector (pNGVL-3). Each treatment consisted of four transfections. One transfection was directly over the tumor site, and three additional treatments were evenly spaced around the circumference of the tumor in a triangle pattern as described (8). Tumor growth was monitored two to three times per week by caliper measurement of two perpendicular tumor diameters. Tumor size was presented as the product of the perpendicular tumor diameters. Mice were killed when the diameter of the tumor reached 10 mm or greater. Mice that had been tumor free for more than 50 days after tumor implantation were challenged with an intradermal injection of 1 × 105 of the parental tumor cells (TS/A) or CT26 tumor cells (murine colon adenocarcinoma), which is also syngeneic to BALB/c mice.
Analysis of ICE mRNA Expression in Tumors by RT-PCR.
Total cellular RNA was extracted from the ICE cDNA-transfected and nontransfected TS/A, LLC, or B16 cells by using a RNeasy Mini Kit (Qiagen, Chatsworth, CA). As a positive control or negative control, expression of mRNA extracted from RAW 264.7 cells or COS-7 cells, respectively, also was tested (25, 26, 36, 37). Three micrograms of total RNA were digested by RQ1 RNase-free DNase (Promega) and were reverse transcribed by using a Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega). Five hundred nanograms of RNA-equivalent cDNA were amplified by PCR under the following conditions: 94°C, 2 min; 57°C, 1 min; and 72°C, 1 min for 35 cycles. The following sequences were used as primers: ICE, (sense) 5′-GTCTTGCCCTCATTATCTGC-3′ and (antisense) 5′-GGTG TTGAAGAGCAGAAAGC-3′; β-actin, (sense) 5′-TGTCCCTGTATGCCTCTGGT-3′ and (antisense) 5′-ACTGTGTTGGCATAGAGGTC-3′. The PCR products of ICE and β-actin are fragments of 516 and 482 bp in length, respectively. The amplified products were electrophoresed on 2.0% agarose gel, and the intensity of DNA bands was quantified by densitometry (National Institutes of Health image 1.61).
mIFN-γ Expression in Vivo.
To analyze IFN-γ production after in vivo gene transfer, blood, gene-gun-treated skin tissue, and splenocytes were obtained 24 hr after the second gene gun treatment. The serum was frozen at −80°C until assayed. Tissue samples were homogenized in an extraction buffer containing 0.1% Triton X-100 and 1 mM Pefabloc, a protease inhibitor (Boehringer Mannheim) and were sonicated and centrifuged. The supernatants also were frozen at −80°C until assayed. Splenocytes freshly isolated from test mice were cultured for 24 hr in the absence of Con A, and the supernatants also were frozen at −80°C until assayed. The quantification of mIFN-γ production in the gene gun-treated skin tissues and serum of test mice and in conditioned medium of cultured splenocytes was performed by ELISA using commercially available purified and biotinylated antibody pairs (PharMingen), streptavidin-horseradish peroxidase conjugate (Zymed), and tetramethylbenzidine substrate (Dako). The lower limit of detection sensitivity was 78 pg/ml. All samples were assayed in duplicates.
In Vivo Depletion of CD4+ and CD8+ T cells and Neutralization of IFN-γ.
The relative contribution of T cell subsets and IFN-γ was evaluated by in vivo antibody inhibition. Anti-CD4 mAb (clone GK1.5) and anti-CD8 mAb (clone 2.43) were administered intraperitoneally at 0.3 mg per injection per mouse on days 5, 9, and 13 after tumor implantation (8). Anti-IFN-γ mAb (clone R4–6A2) was injected intraperitoneally at 0.5 mg per mouse on days 5, 7, 9, 11, and 13. As an isotype antibody, rat IgG (Sigma) was injected at 0.5 mg per mouse with the same schedule as for the anti-IFN-γ mAb.
Cytotoxic Assay.
Spleen cells (8 × 106), derived from BALB/c mice that rejected TS/A tumors after gene therapy and remained tumor-free for at least 50 days, or obtained from TS/A tumor bearing mice 2 weeks after tumor implantation, were co-cultured with irradiated TS/A cells (4 × 105) for 5 days in vitro. TS/A cells labeled with Na251CrO4 (5 × 103/well) were cultured in a total volume of 200 μl with effector cells in 96 round-bottomed well plate. After 4 hr of incubation, the supernatant was harvested and counted in a gamma counter, and specific lysis was calculated (8).
Results and Discussion
ICE mRNA Expression in Tumor Cells.
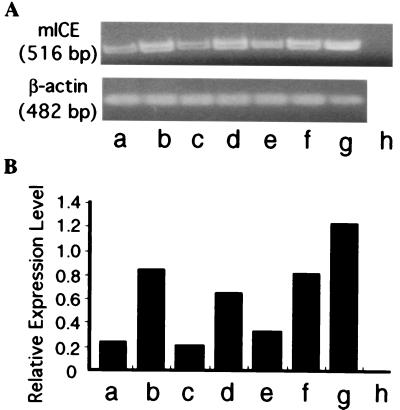
The coexpression of murine pro-IL-18 with murine ICE, but not pro-IL-18 alone, in COS cells results in the secretion of bioactive IL-18 protein (25). Cleavage of pro-IL-18 by ICE results in secretion of the active form of IL-18 from the cells (26). We postulated that the expression of ICE is essential to generate transgenic IL-18 from the test cells and analyzed the expression of ICE mRNA of tumor cells. Several tumor cell lines, including TS/A, LLC, and B16, had basal levels of ICE mRNA expression (Fig. 1A). In comparison, the transfection of these cells with ICE cDNA resulted in a 3- to 4-fold increase in ICE mRNA expression (Fig. 1B). As a positive control, RAW264.7 cells were used in these assays (36, 37), and COS-7 cells were used as a negative control (25, 26).
Figure 1.
(A) ICE mRNA expression of the tumor cell lines by RT-PCR. (B) Densitometric analysis of ICE mRNA expression. (A) The cDNA samples obtained by RT-PCR for mICE and β-actin were loaded on the 2% agarose gel. (B) Relative expression level means the ratio obtained from the density of band of ICE mRNA expression standardized by that of β-actin mRNA expression. The representative data of three separate experiments are shown. a, TS/A; b, TS/A transfected with ICE cDNA; c, LLC; d, LLC transfected with ICE cDNA; e, B16; f, B16 transfected with ICE cDNA; g, RAW 264.7; h, COS-7 cells.
Induction of IFN-γ by Gene Transfer of IL-12, pro-IL-18, and/or ICE.
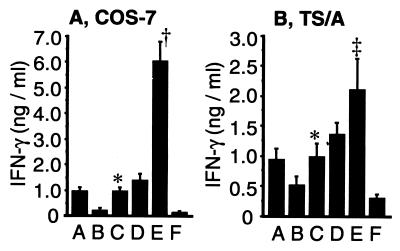
We hypothesized that expression of endogenous ICE in tumor cells may result in cleavage of the transgenic pro-IL-18. To evaluate this hypothesis, we transfected COS-7 cells (ICE-negative) and TS/A (ICE-positive) cells with pro-IL-18 cDNA alone or in combination with an ICE cDNA expression vectors. The ability of these cells to stimulate IFN-γ production in Con A-primed murine splenocytes was used as a bioassay to measure the relative levels of functional IL-18 protein (18, 25, 26). Fig. 2 shows that IFN-γ level induced by transfection with pro-IL-18 and ICE was considerably higher than that by pro-IL-18 alone in both COS-7 and TS/A cells. Specifically, IFN-γ production for pro-IL-18/ICE versus pro-IL-18 alone was 977 ± 135 versus 247 ± 57 pg/ml, P < 0.05, for COS-7 cells, and was 1,007 ± 214 versus 537 ± 129 pg/ml, P < 0.05, for TS/A cells. As expected, the cotransfection of ICE and pro-IL-18 cDNA was superior to pro-IL-18 cDNA alone and resulted in enhanced bioactivity of IL-18 in the ICE negative cell line COS-7 (25). Both the low level of endogenous ICE in TS/A cells and subsequent low level of IFN-γ production, however, were significantly enhanced by ICE cDNA cotransfection. Thus, dual transfection of pro-IL-18 and ICE cDNA results in secretion of more bioactive IL-18 protein than that of pro-IL-18 cDNA alone.
Figure 2.
Induction of IFN-γ in splenocytes in vitro by cytokine protein secreted from transfected COS-7 (A) and TS/A (B) cells. IL-18 bioassay (stimulation of IFN-γ release) was carried out as described in Materials and Methods. Means ± SD were calculated from triplicates. *, P < 0.05 versus pro-IL-18 alone in both cells; †, P < 0.0001 versus the other groups in COS-7 cells; ‡, P < 0.005 versus the other groups in TS/A cells. A, IL-12 cDNA; B, pro-IL-18 cDNA; C, pro-IL-18/ICE cDNA; D, IL-12/pro-IL-18 cDNA; E, IL-12/pro-IL-18/ICE cDNA; F, control plasmid (pNGVL-3).
We also evaluated whether the addition of an expression vector encoding mIL-12 could further enhance IFN-γ production in vitro. The combination of IL-12, pro-IL-18, and ICE resulted in the highest level of IFN-γ release from Con A-primed splenocytes among all groups in COS-7 and TS/A cells (Fig. 2). IFN-γ levels induced by IL-12, pro-IL-18, and ICE in combination were 6,070 ± 713 pg/ml and 2,113 ± 508 pg/ml in COS-7 and TS/A cultures, respectively. In contrast, the IFN-γ level induced by pro-IL-18 and IL-12 together was not statistically different from that of treatment with IL-12 alone. Taken together, in terms of IFN-γ induction, IL-12 was synergistic with the IL-18 produced from ICE-cleaved pro-IL-18 but not with pro-IL-18 alone. Similar results were obtained in transfection studies performed with LLC and B16 cells (data not shown).
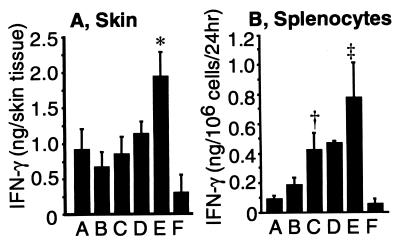
To investigate whether IFN-γ levels could be augmented in vivo after the transfer of IL-12, pro-IL-18, and ICE cDNA, IFN-γ production in the gene gun-treated skin tissues and serum of test mice and in conditioned medium of cultured splenocytes were compared among various treatment groups. The IFN-γ level produced from the skin in the group of IL-12/pro-IL-18/ICE was 1,949 ± 332 pg per tissue site, which was the highest among all tested groups (Fig. 3A). The level of IFN-γ in the serum of test mice for all groups was not detectable within the sensitivity limit of our ELISA assay (=78 pg/ml). When splenocytes freshly isolated from test mice were cultured for 24 hr in the absence of Con A, the level of IFN-γ secretion was greatest in the group that was treated with the set of IL-12, pro-IL-18, and ICE cDNA in combination (781 pg/106 splenocytes/24 hr; Fig. 3B). This result suggests that splenocytes can be significantly activated after in vivo gene delivery of IL-12, pro-IL-18, and ICE and that gene gun-mediated skin transfection with IL-12, pro-IL-18, and ICE cDNA may be capable of stimulating immune cells systemically as well as locally.
Figure 3.
IFN-γ production in the treated skin tissue (A) and from splenocytes (B) after gene gun treatment. (A) Skin overlying TS/A tumor was excised after the second gene gun treatment, and IFN-γ in the skin tissue lysate was measured by ELISA. *, P < 0.05 versus all other treatment groups. Mean ± SD are shown for four mice per group. (B) Splenocytes (2 × 106) were isolated after the second gene gun treatment and were cultured in 2 ml of RPMI medium 1640 for 24 hr. †, P < 0.05 versus pro-IL-18 alone; ‡, P < 0.05 versus all other groups. Mean ± SD are shown for four mice per group. A, IL-12 cDNA; B, pro-IL-18 cDNA; C, pro-IL-18/ICE cDNA; D, IL-12/pro-IL-18 cDNA; E, IL-12/pro-IL-18/ICE cDNA; F, control plasmid (pNGVL-3).
It is also noteworthy that we show that the triple gene set of IL-12, pro-IL-18, and ICE in combination can be successfully delivered in vitro and in vivo by particle-mediated gene transfer with a gene gun device. In addition, in terms of induction of IFN-γ, the finding that transfection of pro-IL-18 and ICE cDNA is superior to that of pro-IL-18 cDNA alone suggests that the two testing genes were concomitantly expressed in the same cell because cleavage of pro-IL-18 by ICE requires the expression of both transgenes within the same cell (25, 26). Thus, particle-mediated gene transfer effectively results in the expression of at least two or perhaps three transgenes in the same cells both in vitro and in vivo. Theoretically, this technique enables multiple gene transfer and expression in the same cells by coprecipitation of multiple species of DNA molecules onto the same gold beads (34, 38).
Antitumor Effect of Gene Gun Treatment with IL-12, pro-IL-18, and ICE cDNA.
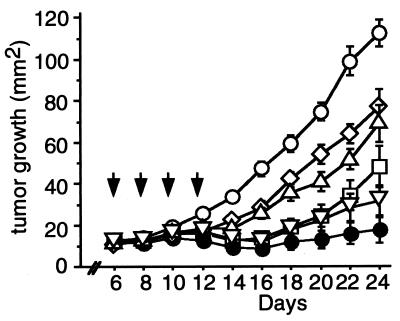
The highest level of IFN-γ induction via the combination of IL-12, IL-18, and ICE treatment among all tested sets, in vitro and in vivo, led us to test whether this combination also could result in regression of established TS/A tumors. The combination gene therapy of IL-12, pro-IL-18, and ICE elicited the most marked suppression of tumor growth among all groups (Fig. 4). Even more important, a complete regression was observed on day 50 in 11 of 22 tested mice (50.0%) in the IL-12/pro-IL-18/ICE combinatorial gene therapy group, and this rate was 2- to 3-fold higher than that obtained from treatment with IL-12 alone or IL-12/pro-IL-18 (Table 1). These results further confirm that the combination of IL-12/pro-IL-18/ICE in this gene therapy strategy can confer a much more efficacious antitumor activity than that from either the IL-12 gene alone or IL-12/pro-IL-18 genes. This is the first evidence that a combinatorial direct treatment with IL-12 and IL-18 cDNA can result in complete eradication of established and relatively large tumors. Previous reports demonstrated efficacy of this combination of genes in a vaccination model (31). The combination of IL-18 and IL-12 protein therapy in a murine model also has been reported. Although tumor regression was noted, the treatment-related toxicity was very high (30). In contrast, no adverse effect was observed in treated mice in our experiments using the combination gene therapy protocol with a gene gun. This suggests that gene gun-mediated local delivery of IL-12/pro-IL-18/ICE genes may be clinically desirable and safe as a strategy for local cancer therapy.
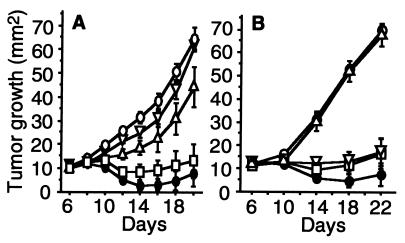
Figure 4.
TS/A growth after in vivo gene gun treatment. The protocol of gene gun treatment is described in Materials and Methods. Arrows indicate the days when gene gun treatments were performed. Mean tumor size ± SEM are shown for 15 mice per each group. A statistically significant difference in suppression of the tumor growth was observed in the group treated with IL-12/pro-IL-18/ICE, compared with that from pro-IL-18 alone (P < 0.05 on day 12 and P < 0.0001 on days 14–24), from pro-IL-18/ICE (P < 0.001 on day 14 and P < 0.0001 on days 16–24), from IL-12 alone (P < 0.05 on day 22 and P < 0.01 on day 24), and from the control plasmid (P < 0.0001 on days 12–24). □, IL-12 cDNA; ⋄, pro-IL-18 cDNA; ▵, pro-IL-18/ICE cDNA; ▿, IL-12/pro-IL-18 cDNA; ●, IL-12/pro-IL-18/ICE cDNA; ○, control plasmid (pNGVL-3).
Table 1.
Complete regression of TS/A tumor after gene gun treatment
| Treatment | Mice with complete regression*/total | Percent |
|---|---|---|
| IL-12 | 4/23 | 17.4† |
| pro-IL-18 | 0/15 | 0‡ |
| pro-IL-18/ICE | 0/15 | 0‡ |
| IL-12/pro-IL-18 | 6/23 | 26.1§ |
| IL-12/pro-IL-18/ICE | 11/22 | 50.0 |
| Control (pNGVL-3) | 0/23 | 0‡ |
Treatment protocol was same as that of Fig. 5 (in IL-12, IL-12/pro-IL-18, IL-12/pro-IL-18/ICE, and control, seven to eight additional mice were evaluated).
The number of mice in which complete regression of established TS/A tumor was observed.
†P < 0.05 vs. IL-12/pro-IL-18/ICE.
‡P < 0.0001 vs. IL-12/pro-IL-18/ICE.
§P = 0.089 vs. IL-12/pro-IL-18/ICE.
Results from our present study also demonstrate that IL-12 gene therapy can induce substantially more potent antitumor effects on the established tumor than the IL-18 gene therapy (Fig. 4, IL-12 alone versus pro-IL-18 alone: P < 0.005 on day 14 to 22, and P < 0.05 on day 24; IL-12 versus pro-IL-18/ICE: P < 0.05 on day 14 to 22). In TS/A tumor-bearing mice receiving gene gun treatment of IL-12 cDNA, 17.4% of mice showed a complete regression of the established tumors. In contrast, IL-18 gene therapy as administered here has failed to eradicate tumors completely, and all IL-18 cDNA treated mice eventually died from progression of testing tumors (Table 1). Others also have reported that the antitumor effects of SCK cells (murine mammary adenocarcinoma) expressing IL-12 were more striking than those of SCK cells expressing IL-18 in tumor protection model (31).
Importantly, we also showed in this study that the potent antitumor effects induced by IL-12/pro-IL-18/ICE cDNA were in fact reflected by the prolongation of survival time in treated animals. When mice whose tumor did not exceed 100 mm2 in size were defined as survivors, in the groups of mice receiving gene therapy with IL-12/pro-IL-18/ICE, pro-IL-18/IL-12, and IL-12 alone, 59.1, 30.4, and 17.4% of mice, respectively, were found to survive on day 50. There was a statistically significant difference between the combination of IL-12/pro-IL-18/ICE and the rest of the treatment groups (e.g., IL-12/pro-IL-18/ICE versus IL-12 alone, χ2 = 7.98, P = 0.0047). The P value of the IL-12/pro-IL-18 versus the IL-12/pro-IL-18/ICE group was 0.0739 (χ2 = 3.194), which was close to the P < 0.05 significance level (data not shown).
Involvement of CD8+ T Cells and IFN-γ in Tumor Regression After the Combinatorial Gene Therapy of IL-12, pro-IL-18, and ICE.
To determine the mechanism of cytokine-mediated tumor regression, in vivo depletion experiments were performed by injecting TS/A tumor-bearing mice with anti-CD4+, anti-CD8+, or anti-IFN-γ mAb (Fig. 5). Mice treated with IL-12/pro-IL-18/ICE and both anti-CD4+ and CD8+ mAbs were found to develop tumors with kinetics similar to that of the control plasmid group (Fig. 5A). In vivo depletion of CD8+ T cells, but not of CD4+ T cells, completely abrogated the antitumor effect of combination therapy (Fig. 5B). These results suggest that tumor regression induced by this combinatorial therapy requires CD8+ T cells. This finding is similar to the results of antibody depletion experiments for animals receiving IL-12 gene therapy alone (4, 8). Micallef et al. demonstrated that the effector cells responsible for antitumor effects of pretreatment with rIL-18 were NK cells in the initial phase, both CD4+ and CD8+ T cells in the second phase (between day 9 and 15), and CD4+ T cells for the long term immunological memory (27, 28). Our results indicate that the combination therapy of IL-12 and IL-18 can cause antitumor effects primarily via activation of CD8+ T cells.
Figure 5.
Effects of in vivo depletion of CD4+ and CD8+ subsets of T cells and neutralization of IFN-γ on the antitumor response induced by combinatorial gene therapy with IL-12, pro-IL-18, and ICE. (A) Mixture of anti-CD4 and anti-CD8 mAb, or anti-IFN-γ mAb were injected intraperitoneally. Mean tumor size ± SEM are shown for five mice per group. ▿, anti-CD4 and anti-CD8 mAb; ▵, anti-IFN-γ mAb; ●, no mAb; □, rat IgG. All above groups (▿, ▵, ●, and □) received IL-12/pro-IL-18/ICE cDNA treatment. ○, control plasmid (pNGVL-3) and no mAb. (B) Anti-CD4 and anti-CD8 mAb were administered separately. Mean tumor size ± SEM are shown for six mice per group. ▿, anti-CD4 mAb; ▵, anti-CD8 mAb; ●, no mAb; □, rat IgG. All above groups (▿, ▵, ●, and □) received IL-12/pro-IL-18/ICE cDNA treatment. ○, control plasmid (pNGVL-3) and no mAb.
In vivo neutralization of IFN-γ proteins with anti-IFN-γ mAb also inhibited the antitumor activity against the TS/A tumor induced by IL-12/pro-IL-18/ICE cDNA, although this inhibitory effect was not complete (Fig. 5A). This result suggests that IFN-γ is, in part, responsible for the antitumor efficacy of the combination gene therapy of IL-12/pro-IL-18/ICE. The lack of complete abrogation of antitumor efficacy in animals treated with anti-IFN-γ Ab may be explained by persistent local IFN-γ, even in the mice treated with anti-IFN-γ Ab. Moreover, it is likely that IFN-γ may be an important but not necessarily an essential intermediate in this antitumor cascade. This is supported by studies that show that the use of the IFN-γ gene in similar tumor treatment models has not resulted in tumor regression (39), implying that IL-12 and this combination gene therapy can have immunologically mediated antitumor effects other than IFN-γ release. In contrast, Coughlin et al. have demonstrated that the synergistic antitumor effect of IL-12 and IL-18 require the endogenous production of IFN-γ in the vaccination model (31). Others also have reported that IFN-γ is one of the critical factors needed to mediate the antitumor efficacy of IL-18 (29). The observed differences in the role of IFN-γ in IL-12 or in combination with IL-18 gene-mediated antitumor activity might be attributable to the different tumor models applied.
CTL Activity Induced by Gene Gun Delivery of Cytokine cDNAs.
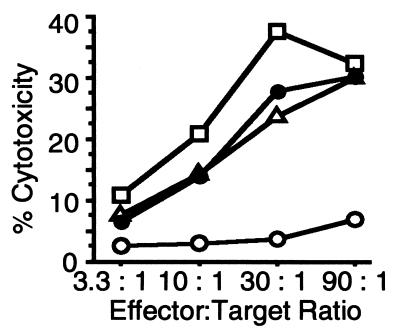
To further evaluate whether the mice that showed complete regression of tumor after in vivo gene transfer were able to develop antitumor immunity, splenocytes collected from mice with complete regression in each treatment group were assayed for CTL activity. Splenocytes from IL-12 alone, IL-12/pro-IL-18, and IL-12/pro-IL-18/ICE gene-treated mice generated similar levels of CTL activity, which were 4- to 6-fold higher than those from control tumor-bearing mice (P < 0.01, Fig. 6).
Figure 6.
Induction of CTL activities in mice with complete regression was observed after gene therapy with IL-12, IL-12/pro-IL-18, or IL-12/pro-IL-18/ICE. The representative data of two independent experiments are shown. □, IL-12 cDNA; ▵, IL-12/pro-IL-18 cDNA; ●, IL-12/pro-IL-18/ICE cDNA; ○, control plasmid (pNGVL-3).
Immunological Memory in Mice After Gene Therapy of IL-12, pro-IL-18, and ICE.
We evaluated whether the mice that showed complete regression of TS/A tumors after gene therapy of IL-12/pro-IL-18/ICE could develop tumor-specific immunity. Six BALB/c mice with complete regression of TS/A tumor after in vivo gene gun treatment with IL-12/pro-IL-18/ICE cDNA were rechallenged on day 50 with 1 × 105 TS/A cells. All mice rejected TS/A tumors and were tumor-free for an additional 1 month. Then, these mice were rechallenged again with 1 × 105 of both TS/A cells and CT26 cells (syngeneic to BALB/c mice) on the right and the left side of abdomen, respectively. All mice rejected a second challenge with TS/A tumor cells but developed CT26 tumors (data not shown). Taken together with the data of the cytotoxic assay (Fig. 6), our results suggest that gene gun treatment with combination of IL-12, pro-IL-18, and ICE in tumor-bearing mice can effectively result in the CTL activation and the development of tumor-specific immunological memory.
In summary, gene gun-mediated transfer of expression vectors for IL-12, pro-IL-18, and ICE cDNA can confer a synergistic induction of IFN-γ in vitro and in vivo. Combination of these three genes can effectively result in complete regression of TS/A tumors, the effect of which is superior to either IL-12 or pro-IL-18 cDNA alone. This antitumor response can be completely blocked by CD8+ T cell depletion and can be partially abrogated by antibodies to IFN-γ. In addition, this combination gene therapy can induce tumor-specific immunological memory. This is a pilot study on the successful use of a triple gene combination for cancer therapy using nonviral vectors. These findings suggest that combination gene therapy of IL-12, IL-18, and ICE cDNA may provide a potential application for cancer gene immunotherapy, and the current gene gun delivery approach may provide a new methodology for effective and functional delivery of multiple, candidate therapeutic genes for experimental and potential clinical applications.
Acknowledgments
This study was supported by the U.S. Army Medical Research Acquisition Activity Grant under DAMD 17-96-2-6017 funded to Dr. N. S. Yang and A. L. Rakhmilevich.
Abbreviations
- ICE
interleukin-1β converting enzyme
- CTL
cytotoxic T lymphocyte
- LLC
Lewis lung carcinoma
- RT
reverse transcription
- m
murine
References
- 1.Dilloo D, Bacon K, Holden W, Zhong W, Burdach S, Zlotnik A, Brenner M. Nat Med. 1996;2:1090–1095. doi: 10.1038/nm1096-1090. [DOI] [PubMed] [Google Scholar]
- 2.Roth J A, Cristiano R J. J Natl Cancer Inst. 1997;89:21–39. doi: 10.1093/jnci/89.1.21. [DOI] [PubMed] [Google Scholar]
- 3.Gilboa E. Semin Oncol. 1996;23:101–107. [PubMed] [Google Scholar]
- 4.Brunda M J, Luistro L, Warrier R R, Wright R B, Hubbard B R, Murphy M, Wolf S F, Gately M K. J Exp Med. 1993;178:1223–1230. doi: 10.1084/jem.178.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahara H, Zeh H J, III, Storkus W J, Pappo I, Watkins S C, Gubler U, Wolf S F, Robbins P D, Lotze M T. Cancer Res. 1994;54:182–189. [PubMed] [Google Scholar]
- 6.Nastala C L, Edington H D, Mckinnery T G, Tahara H, Nalesnik M A, Brunda M J, Gately M K, Wolf S F, Schreiber R D, Storkus W J, et al. J Immunol. 1994;153:1697–1706. [PubMed] [Google Scholar]
- 7.Tannenbaum C S, Wicker N, Armstrong D, Tubbs R, Finke J, Bukowski R M, Hamilton T A. J Immunol. 1996;156:693–699. [PubMed] [Google Scholar]
- 8.Rakhmilevich A L, Turner J, Ford M J, McCabe D, Sun W H, Sondel P M, Grota K, Yang N S. Proc Natl Acad Sci USA. 1996;93:6291–6296. doi: 10.1073/pnas.93.13.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manetti R, Parronchi P, Giudizi M G, Piccinni M P, Maggi E, Trinchieri G, Romagnanim S. J Exp Med. 1993;177:1199–1204. doi: 10.1084/jem.177.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh C S, Macatonia S E, Tripp C S, Wolf S F, O’Garra A, Murphy K. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 11.Martinotti A, Stoppacciaro A, Vagliani M, Melani C, Spreafico F, Wysocka M, Parmiani G, Trinchieri G, Colombo M P. Eur J Immunol. 1995;25:137–146. doi: 10.1002/eji.1830250124. [DOI] [PubMed] [Google Scholar]
- 12.Tsung K, Meko J B, Peplinsky G R, Tsung Y L, Norton J A. J Immunol. 1997;158:3359–3365. [PubMed] [Google Scholar]
- 13.Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 14.Brunda M J, Luistro L, Hendrzak J A, Fountoulakis M, Garotta G, Gately M K. J Immunother. 1995;17:71–77. doi: 10.1097/00002371-199502000-00001. [DOI] [PubMed] [Google Scholar]
- 15.Angiolillo A J, Sgadari C, Taub D D, Liao F, Farberm J M, Maheshwari S, Kleiman H K, Reaman G H, Tosato G. J Exp Med. 1995;182:155–162. doi: 10.1084/jem.182.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sgadari C, Angiolillo A J, Tosato G. Blood. 1996;87:3877–3882. [PubMed] [Google Scholar]
- 17.Okamura H, Nagata K, Komatsu T, Tanimoto T, Nukata Y, Tanabe F, Akita K, Torigoe K, Okura T, Fukuda S, et al. Infect Immun. 1995;63:3966–3972. doi: 10.1128/iai.63.10.3966-3972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamura H, Tsutsui H, Komatsu T, Yutsudo M, Hakura A, Tanimoto T, Torigoe K, Okura T, Nukata Y, Hattori K, et al. Nature (London) 1995;378:88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 19.Kohno K, Kataoka J, Ohtsuki T, Suemoto Y, Okamoto I, Usui M, Ikeda M, Kurimoto M. J Immunol. 1997;158:1541–1550. [PubMed] [Google Scholar]
- 20.Ushio S, Namba M, Okura T, Hattori K, Nukada Y, Akita K, Tanabe F, Konishi K, Micallef M, Fujii M, et al. J Immunol. 1996;156:4274–4279. [PubMed] [Google Scholar]
- 21.Micallef M, Ohtsuki T, Kohno K, Tanabe F, Ushio S, Namba M, Tanimoto T, Torigoe K, Fujii M, Ikeda M, et al. Eur J Immunol. 1996;26:1647–1651. doi: 10.1002/eji.1830260736. [DOI] [PubMed] [Google Scholar]
- 22.Dao T, Ohashi K, Kayano T, Kurimoto M, Okamura H. Cell Immunol. 1996;173:230–235. doi: 10.1006/cimm.1996.0272. [DOI] [PubMed] [Google Scholar]
- 23.Tsutsui H, Nakanishi K, Matsui K, Higashino K, Okamura H, Miyazawa Y, Kaneda K. J Immunol. 1996;157:3967–3973. [PubMed] [Google Scholar]
- 24.Dao T, Mehal W Z, Crispe I N. J Immunol. 1998;61:2217–2222. [PubMed] [Google Scholar]
- 25.Ghayur T, Banerjee S, Hugunin M, Butler D, Herzog L, Carter A, Quintal L, Sekut L, Talanian R, Paskind M, et al. Nature (London) 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 26.Gu Y, Kuida K, Tsutsui H, Ku G, Hsiao K, Fleming M A, Hayashi N, Higashino K, Okamura H, Nakanishi K, et al. Science. 1997;275:206–209. doi: 10.1126/science.275.5297.206. [DOI] [PubMed] [Google Scholar]
- 27.Micallef M J, Tanimoto T, Kohno K, Ikeda M, Kurimoto M. Cancer Res. 1997;57:4557–4563. [PubMed] [Google Scholar]
- 28.Micallef M J, Yoshida K, Kawai S, Hanaya T, Kohno K, Arai S, Tanimoto T, Torigoe K, Fujii M, Ikeda M, et al. Cancer Immunol Immunother. 1997;43:361–367. doi: 10.1007/s002620050345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan J, Crucian B E, Chang A E, Aruga E, Aruga A, Dovhey S E, Tanigawa K, Yu H. J Immunother. 1997;21:48–55. doi: 10.1097/00002371-199801000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Osaki T, Peron J M, Cai Q, Okamura H, Robbins P D, Kurimoto M, Lotze M T, Tahara H. J Immunol. 1998;160:1742–1749. [PubMed] [Google Scholar]
- 31.Coughlin C M, Salhany K E, Wysocka M, Aruga E, Kurzawa H, Chang A E, Hunter C A, Fox J C, Trinchieri G, Lee W M F. J Clin Invest. 1998;101:1441–1452. doi: 10.1172/JCI1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn H J, Mauro S, Tomura M, Mu J, Hamaoka T, Nakanishi K, Clark S, Kurimoto M, Okamura H, Fujisawa H. J Immunol. 1997;159:2125–2131. [PubMed] [Google Scholar]
- 33.Marshall E. Science. 1995;268:1555. [Google Scholar]
- 34.Sun W H, Burkholder J K, Sun J, Culp J, Turner J, Lu X G, Pugh T D, Ershler W B, Yang N S. Proc Natl Acad Sci USA. 1995;92:2889–2893. doi: 10.1073/pnas.92.7.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahvi D M, Burkholder J K, Turner J, Culp J, Malter J S, Sondel P M, Yang N S. Hum Gene Ther. 1996;7:1535–1543. doi: 10.1089/hum.1996.7.13-1535. [DOI] [PubMed] [Google Scholar]
- 36.Casano F J, Rolando A M, Mudgett J S, Molineaux S M. Genomics. 1994;20:474–481. doi: 10.1006/geno.1994.1203. [DOI] [PubMed] [Google Scholar]
- 37.Nett M A, Cerretti D P, Berson D R, Seavitt J, Gilbert D J, Jenkins N A, Copeland N G, Black R A, Chaplin D D. J Immunol. 1992;149:3254–3259. [PubMed] [Google Scholar]
- 38.Albertini M R, Elmer C A, Schell K, Tans K J, King D M, Sheehy M J. Cancer Gene Ther. 1996;3:192–201. [PubMed] [Google Scholar]
- 39.Rakhmilevich A L, Janssen K, Turner J, Culp J, Yang N S. Hum Gene Ther. 1997;8:1303–1311. doi: 10.1089/hum.1997.8.11-1303. [DOI] [PubMed] [Google Scholar]