Abstract
In this study, we establish that cholesterol and sphingolipid associated with hepatitis C virus (HCV) particles are important for virion maturation and infectivity. In a recently developed culture system enabling study of the complete life cycle of HCV, mature virions were enriched with cholesterol as assessed by the molar ratio of cholesterol to phospholipid in virion and cell membranes. Depletion of cholesterol from the virus or hydrolysis of virion-associated sphingomyelin almost completely abolished HCV infectivity. Supplementation of cholesterol-depleted virus with exogenous cholesterol enhanced infectivity to a level equivalent to that of the untreated control. Cholesterol-depleted or sphingomyelin-hydrolyzed virus had markedly defective internalization, but no influence on cell attachment was observed. Significant portions of HCV structural proteins partitioned into cellular detergent-resistant, lipid-raft-like membranes. Combined with the observation that inhibitors of the sphingolipid biosynthetic pathway block virion production, but not RNA accumulation, in a JFH-1 isolate, our findings suggest that alteration of the lipid composition of HCV particles might be a useful approach in the design of anti-HCV therapy.
Hepatitis C virus (HCV) is recognized as a major cause of chronic liver disease, including chronic hepatitis, hepatic steatosis, cirrhosis, and hepatocellular carcinoma. It presently affects approximately 200 million people worldwide (26). HCV is an enveloped positive-strand RNA virus belonging to the Hepacivirus genus of the family Flaviviridae. Its genome of ∼9.6 kb encodes a polyprotein precursor of ∼3,000 residues, and the structural proteins (core, E1, and E2) reside in its N-terminal region.
Little is known about the assembly of HCV and its virion structure, because efficient production of authentic HCV particles has only recently been achieved. Nucleocapsid assembly generally involves oligomerization of the capsid protein and encapsidation of genomic RNA. This process is thought to occur upon interaction of the core protein with viral RNA, and this core-RNA interaction may induce a change from RNA replication to packaging. As with related viruses, the mature HCV virion likely consists of a nucleocapsid and an outer envelope composed of a lipid membrane and envelope proteins. Expression of the structural proteins in mammalian cells has been observed to generate virus-like particles with ultrastructural properties similar to those of HCV virions (5, 29). Packaging of these HCV-like particles into intracellular vesicles as a result of budding from the endoplasmic reticulum (ER) has also been observed (8, 34). However, HCV structural proteins are observed both in the ER and in the Golgi apparatus (45). Moreover, complex N-linked glycans have been detected on the surfaces of HCV particles isolated from patient sera, suggesting that the glycans transit through the Golgi apparatus (44). Interactions between the core and E1/E2 proteins are thought to determine viral morphology and are mediated through a cytoplasmic loop present in the polytopic form of E1 (35). Recently, we and others have identified a unique HCV genotype 2a isolate, JFH-1, that is able to replicate and produce high levels of infectious virus in culture (HCVcc) (54, 56), enabling us to investigate new aspects of the HCV life cycle.
In this study, we examine the importance of cholesterol and sphingolipid in association with the HCV membrane in virion maturation and virus infectivity. Mature HCV particles are rich in cholesterol. Cholesterol depletion or hydrolysis of sphingolipid from HCV particles results in a loss of infectivity. We further demonstrate a requirement for virion-associated cholesterol and sphingolipid for viral entry.
MATERIALS AND METHODS
Cell culture.
The human hepatoma cell line Huh-7, which is permissive to HCV infection, was obtained from Francis V. Chisari (The Scripps Research Institute). Human embryonic kidney 293T cells were cultured in Dulbecco's modified Eagle medium (DMEM)-10% fetal bovine serum. Huh-7 cell lines, which carry subgenomic replicon RNA of either the JFH-1 (20) or the N (11, 17) strain, were cultured as previously described (21, 46).
Reagents.
The primary antibodies used in this study were mouse monoclonal antibodies against vesicular stomatitis virus glycoprotein (VSV-G) (Sigma, St. Louis, MO), HCV E1 (54) and E2 (Biodesign International, Saco, ME), caveolin-2 (New England Biolabs, Beverly, MA), and CD81 (BD Pharmingen, Franklin Lakes, NJ), as well as rabbit polyclonal antibodies against calnexin (Stressgen, Ann Arbor, MI) and HCV core (48). ISP-1/myriocin, cholesterol, and heparinase I were purchased from Sigma, and recombinant Bacillus cereus sphingomyelinase (SMase) was obtained from Higeta Shoyu (Tokyo, Japan). (1R,3R)-N-(3-Hydroxy-1-hydroxymethyl-3-phenylpropyl) dodecanamide (HPA-12), which was synthesized as described elsewhere (24), was a gift from Shu Kobayashi (University of Tokyo).
Plasmids.
pCAE1 and pCAE2 contain HCV cDNAs spanning the E1 region (amino acids 192 to 383) with a FLAG tag at the N terminus and the E2 region (amino acids 384 to 809) with a Myc tag at the N terminus of strain NIHJ1 (1), respectively, under the control of the CAG promoter (38). pCAV340V and pCAV711V consist of the ectodomains of E1 and E2, respectively, with the N-terminal signal sequences, transmembrane domains, and cytoplasmic domains derived from VSV-G, as described elsewhere (50) (see Fig. 4D).
FIG. 4.
Compartmentation of HCV structural proteins within DRM fractions. Lysates of HCVcc-infected cells were either treated with 1% TX-100, either on ice or at 37°C, or left untreated, followed by sucrose gradient centrifugation. (A and B) For each fraction, the amount of core protein was determined by an enzyme-linked immunosorbent assay (A), and E1, calnexin, and caveolin-2 were analyzed by Western blotting (B). The amount of core protein in each lysate (TX-100, 37°C; TX-100, 4°C; Untreated) was assigned the arbitrary value of 100%. M, membrane; NM, nonmembrane; DS, detergent soluble. (C) Lysates of 293T cells expressing HCV E1 or E2 protein were either treated with 1% TX-100, either on ice or at 37°C, or left untreated, followed by discontinuous sucrose gradient centrifugation. Each fraction was concentrated in a Centricon YM-30 filter unit and subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting with antibodies against calnexin, caveolin-2, Myc (E1), or FLAG (E2). (D) (Top) Structures of HCV envelope genes used. Amino acid positions of HCV are indicated. Signal sequence, transmembrane (TM), and cytoplasmic tail (CT) domains of VSV G protein are shown. (Bottom) Cell lysates expressing chimeric HCV E1 or E2 protein were treated with 1% TX-100 on ice or left untreated, followed by discontinuous sucrose gradient centrifugation. It has been reported that VSV-G is not associated with lipid (39). Calnexin, caveolin-2, and chimeric glycoproteins (chimeric E1 and chimeric E2) were analyzed by immunoblotting. Fractions are numbered from 1 to 9 in order from top to bottom (light to heavy).
Virus production.
Plasmid pJFH1, containing full-length cDNA of the JFH-1 isolate, was used to generate HCVcc as described elsewhere (23, 33, 34, 54). pJ6/JFH was obtained from JFH1 by replacement of the 5′ untranslated region to the p7 region (EcoRI-BclI) of J6. In vitro-transcribed RNA from linearized pJFH1 or pJ6/JFH1 was delivered to Huh-7 cells by electroporation. Culture supernatants were collected at 72 h posttransfection, clarified by low-speed centrifugation, passed through a 0.45-mm-pore-size filter, and concentrated using an Amicon Ultra-15 unit (Millipore, Bedford, MA) or by ultracentrifugation (23). Infectious titers, HCV RNA copies, and core protein concentrations of the viral stocks were ∼5 × 103 focus-forming units per ml, ∼1 × 107 copies/ml, and ∼1 × 104 fmol/liter, respectively. HCVcc was isolated by a combination of ultrafiltration, ion-exchange chromatography, heparin affinity chromatography, and sucrose density ultracentrifugation (33; K. Morikawa and T. Wakita, unpublished data). Pseudotyped VSV containing E1 and E2 proteins of the HCV genotype 1a isolate H77c (HCVpv) was generated as previously described (51). Briefly, 293T cells transiently expressing E1 and E2 proteins (strain H77) were infected with VSVdelG-GFP/G, in which the G envelope gene was replaced with green fluorescent protein (GFP) and pseudotyped with VSV-G.
Determination of cholesterol and phospholipid contents of HCVcc and infected cells.
Cellular and viral lipids were extracted from isolated HCVcc and from uninfected and infected Huh-7 cells. Cholesterol content was determined using the cholesterol oxidase method as previously described (14). Total phospholipid content was determined using the method of Rouser et al. (42).
Cholesterol depletion and replacement.
To remove cholesterol from the HCV envelope, stock samples of HCVcc were treated with methyl-β-cyclodextrin (B-CD) in DMEM (Sigma) supplemented with 10% fetal bovine serum (Sigma) and nonessential amino acids (Invitrogen, Carlsbad, CA) for 1 h at 37°C, followed by centrifugation at 100,000 × g for 3 h to form a pellet, which was resuspended in 0.5 ml of the medium. In order to replenish cholesterol, the medium of HCVcc treated with 5 mg/ml B-CD was replaced with DMEM containing various concentrations of exogenous cholesterol (Sigma) and incubated for 1 h, followed by centrifugation to form a pellet. In order to perform HCVcc infection assays, Huh-7 cells were infected with HCVcc, with or without the treatment described above, for 1 h at 37°C and then washed as described above. Viral core protein levels in the cells and in the supernatant were quantified 72 h later using an HCV core enzyme-linked immunosorbent assay (Ortho-Clinical Diagnostics, Tokyo, Japan).
SMase treatment.
HCVcc was treated with SMase at various concentrations in DMEM for 1 h at 37°C and was then centrifuged at 100,000 × g for 3 h to form a pellet, which was resuspended in 0.5 ml of medium for the infection assays.
HCVcc binding and internalization assays.
To monitor binding, cells grown in a 6-well plate were preincubated for 1 h at 4°C, after which B-CD- or SMase-treated HCVcc was bound to the cells for 1 h at 4°C. As a measure of virus internalization, following the virus binding procedure, the cells were warmed to 37°C and maintained for 2 h, after which they were treated with 0.25% trypsin for 10 min at 37°C. Huh7-25, a CD81-negative Huh-7 subclone (3), was used to ensure removal of surface-bound virus by trypsin treatment. For both the binding and internalization assays, the resulting cells, as described above, were washed with ice-cold phosphate-buffered saline, followed by lysis with TRIzol reagent (Invitrogen). Cell-associated virus was quantified by measuring the amount of HCV RNA in the cell lysate by the real-time reverse transcription-PCR method (2, 34). Cells were treated with heparinase as previously described (33).
HCV replication assay in HCVcc-infected or replicon cells.
HCV subgenomic replicon cells or cells infected with HCVcc were treated with various concentrations of inhibitors for 72 h. Total RNA was isolated from replicon cells using TRIzol reagent (Invitrogen), followed by quantification of HCV RNA by real-time reverse transcription-PCR as previously described (2, 34). Levels of core protein in the culture supernatants of HCVcc-infected cells were tested as described above.
Detection of cholesterol content of HCVcc.
For [3H]cholesterol labeling of viruses, HCVcc-infected or uninfected cells were incubated with 50 mCi of [1α,2α-3H]cholesterol in DMEM for 24 h. Culture supernatants of the cells were incubated in the presence or absence of B-CD at 5 mg/ml for 1 h at 37°C, followed by ultracentrifugation on a 60% sucrose cushion. The virus-containing fractions and corresponding fractions from an uninfected culture were lysed in the buffer containing 1% Triton X-100 (TX-100), and radioactivity was quantified by scintillation counting. Radioactivities (in counts per minute) of HCVcc samples were determined by subtracting the radioactivity of uninfected cells from that of HCVcc-infected cells.
Metabolic labeling analysis of sphingolipid content.
After 2 h of incubation with [14C]serine (0.5 mCi/ml) in Opti-MEM (Invitrogen), the cells were lysed with 0.1% sodium dodecyl sulfate, and total lipid was extracted with chloroform-methanol (1:2, vol/vol). The extracts were spotted onto silica gel 60 plates (Merck, Darmstadt, Germany) and chromatographed with methyl acetate-1-propanol-chloroform-methanol-0.25% KCl (25:25:25:10:9, vol/vol). Radioactive spots were quantitatively detected by BAS 2000 (Fuji Film, Japan).
Membrane flotation assay.
The membrane flotation assay was performed as previously described (46).
RESULTS
Critical role of virion-associated cholesterol.
A role of virion-associated cholesterol in infectivity has been demonstrated for several enveloped viruses (4). However, little is known about the role of lipids associated with the virions of flaviviruses, including HCV, and their contribution to the viral life cycle. To determine the lipid composition of mature HCV virions, we extracted total lipid from HCVcc (JFH-1 and chimeric J6/JFH-1) prepared from the culture supernatants of cells infected with HCV, as well as the total cellular membrane fractions of uninfected and infected Huh-7 cells. The cholesterol and phospholipid contents were quantified, because these are the two major lipid constituents of biological membranes. The cholesterol-to-phospholipid molar ratio, which is known as a parameter of membrane viscosity (47), was significantly higher in virus samples (1.29 and 1.26 for JFH-1 and J6/JFH-1, respectively) than in cell membrane samples (0.40 and 0.42 for JFH-1-infected and uninfected cells, respectively) (Table 1). The ratios in viral samples were similar to or greater than those in mammalian plasma membranes, where most cellular cholesterol is found. Minimal contamination of the viral samples with extracellular microvesicles likely occurred, since only a small amount of lipid was detected in a sample prepared from the culture medium of uninfected cells (data not shown). Thus, it is likely that HCV virions are enriched with cholesterol during assembly and maturation.
TABLE 1.
Cholesterol and phospholipid contents of HCVcc and cells
| Cell type or virus | Content (nmol/mg of protein)a
|
Chol/PL ratio | |
|---|---|---|---|
| Chol | PL | ||
| Cells | |||
| Uninfected | 105.9 ± 10.4 | 253.2 ± 10.6 | 0.42 |
| JFH-1 infected | 116.5 ± 10.0 | 292.0 ± 18.4 | 0.40 |
| Virus | |||
| JFH-1 | 43.6 ± 2.4 | 33.8 ± 1.8 | 1.29 |
| J6/JFH-1b | 28.7 ± 4.8 | 22.7 ± 2.9 | 1.26 |
Data are averages of three independent measurements ± standard deviations. Chol, cholesterol; PL, phospholipids.
J6/JFH1 virus was produced from the pJ6/N2X-JFH1 construct and has structural proteins from the J6CF strain.
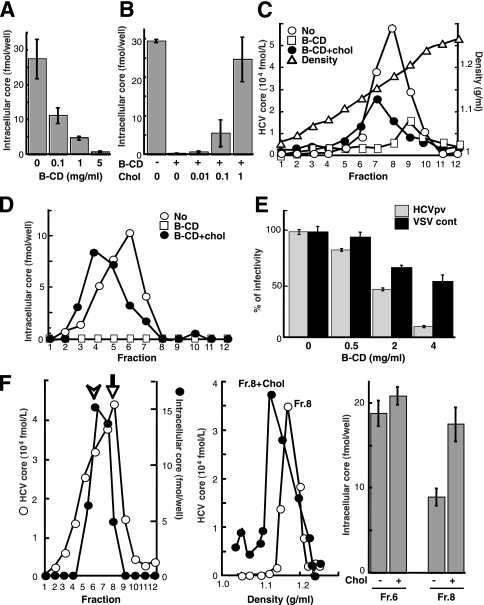
To investigate a potential role for the particular lipid composition of HCV particles, HCVcc was treated with increasing concentrations (0.1 to 5 mg/ml) of B-CD, which is known to extract cholesterol from membranes (40). The viral samples were then used to inoculate Huh-7 cells after removal of B-CD by ultracentrifugation. Infectivity was evaluated by quantifying the viral core protein in cells at 72 h postinfection (p.i.). Using an immunoassay that provides results indicative of HCV infectivity (25), we also confirmed a good correlation between the core level and infectious titers (data not shown). As shown in Fig. 1A, core protein levels following B-CD treatment at 0.1, 1, or 5 mg/ml were reduced by 60, 83, or 98%, respectively, from the levels with the untreated virus. The cholesterol level of HCVcc treated with 5 mg/ml B-CD was found to be ∼50% of that of untreated virions (Table 2).
FIG. 1.
Role of HCV-associated cholesterol in infection. (A) Effect of cholesterol depletion on HCV infectivity. HCVcc particles (∼2 fmol of the core protein) were treated with B-CD at 0.1, 1, and 5 mg/ml for 1 h at 37°C. After removal of B-CD, Huh-7 cells were infected with the treated virus particles, after which the core protein content of infected cells at 72 h p.i. was determined as an indicator of infectivity, as previously established (24). (B) Effect of cholesterol replenishment on infectivity. After treatment with 5 mg/ml B-CD, virus was treated either with medium alone or with medium containing exogenous cholesterol for 1 h at 37°C. (C) Effect of cholesterol depletion and replenishment on density gradient profiles of the viral particles. The HCVcc treated with 5 mg/ml B-CD was replenished with exogenous cholesterol (1 mM) and then separated by 10-to-60% sucrose gradient ultracentrifugation. The core protein in each fraction was measured. The density of each fraction was determined by refractive index measurement. (D) Effects of cholesterol depletion and replenishment on viral infectivity. Each fraction (see panel C) was infected, and then the core proteins in the cells were measured at 72 h p.i. (E) Effect of cholesterol depletion on the infectivity of HCVpv (genotype 1a) (shaded bars) or the control, VSVdelG-GFP/G (solid bars). The viruses were preincubated with B-CD for 1 h at 37°C before infection. (F) (Left) The culture medium from HCVcc-producing cells was fractionated as described above. For each fraction, the amounts of core and intracellular core (infectivity) are plotted. Peaks of the core (arrow) and infectivity (arrowhead) are indicated. (Center) An aliquot of fraction 8 (peak of the core) was treated with 1 mM cholesterol for 1 h at 37°C. The resultant aliquot and an untreated aliquot of the fraction were subjected to sucrose gradient ultracentrifugation. The core in each fraction was plotted. (Right) The infectivities of fractions (Fr.) 6 and 8 (see the left panel) with or without cholesterol treatment were determined as shown above. Data are means from four independent experiments. Error bars, standard deviations.
TABLE 2.
Depletion of virion-associated cholesterol by B-CD
| Treatment | Radioactivity (cpm) of HCVcca
|
Avg (%b) | |
|---|---|---|---|
| Expt 1 | Expt 2 | ||
| None | 5,327 | 5,573 | 5,450 (100) |
| B-CD (5 mg/ml) | 3,643 | 1,646 | 2,644 (48.5) |
Determined by subtracting the radioactivity of uninfected cells from that of HCVcc-infected cells in two experiments.
Percentage of the radioactivity of the untreated sample.
To demonstrate that the reduced infection efficiency of B-CD-treated virus was caused by the reduced cholesterol content of the viral envelope, we attempted to reverse the inhibitory effect by adding exogenous cholesterol. Following treatment of HCVcc with 5 mg/ml B-CD, the drug was washed out, and increasing concentrations of cholesterol were added in an attempt to reconstitute the normal virion cholesterol content. The addition of 1 mM cholesterol completely reversed the virus infectivity (Fig. 1B). After cholesterol was replenished, the viral RNA was restored to a level similar to that in the untreated control.
To investigate the effect of cholesterol on the density of infectious HCV virions, B-CD-pretreated or untreated viral samples, as well as cholesterol-replenished treated viral samples, were subjected to sucrose density gradient centrifugation (Fig. 1C). The density of HCVcc core protein at its peak concentration in untreated virus samples was ∼1.17 g/ml. When virion-associated cholesterol was removed by B-CD, the density of HCVcc core protein at its peak concentration was shifted to 1.20 g/ml. Addition of exogenous cholesterol to this cholesterol-depleted sample restored a lower-density fraction (1.15 g/ml). Figure 1D illustrates the infectivity of each gradient fraction. Untreated virus had maximum infectivity at ∼1.13 g/ml (fraction 6), while, as expected, fractions from B-CD-treated viral samples exhibited minimal to no infectivity. Replenishment of depleted virus with cholesterol returned infectivity to untreated-control levels, and cholesterol-replenished virus had a buoyant density of ∼1.07 g/ml (fraction 4), suggesting that HCV-associated cholesterol is crucial for viral infectivity and that the effect of a cholesterol-depleting drug is reversible. We further observed that B-CD treatment of a pseudotyped VSV containing the E1 and E2 proteins of the HCV genotype 1a isolate H77c (HCVpv) resulted in a progressive loss of infectivity, while B-CD had significantly less impact on the infectivity of the control virus VSVdelG-GFP/G (Fig. 1E).
The results described above raise the possibility that the infectivity of HCV virions with relatively low levels of incorporated cholesterol might be enhanced by supplementation with exogenous cholesterol. Density gradient fractions of culture supernatants collected from HCV-infected cells were analyzed with regard to the presence of core protein and infectivity (Fig. 1F, left). As indicated above, maximum infectivity was obtained with fraction 6 (1.13 g/ml). In contrast, a major fraction of core protein banded at a higher density (1.17 g/ml) in fraction 8. We hypothesized that fraction 8 contains lipids at lower levels than those in fraction 6. However, quantification of lipids, including cholesterol, in the fractions obtained failed, presumably due to a low sensitivity of detection. Thus, to extend our findings on the involvement of cholesterol, we added exogenous cholesterol to fraction 8, followed by ultrafiltration to remove unincorporated cholesterol. A subsequent density gradient profile demonstrated a shift in the core protein peak to 1.13 g/ml (Fig. 1F, center). A concomitant increase in the infectivity of the fraction, approaching that of untreated fraction 6, was observed (Fig. 1F, right). In contrast, supplementation of fraction 6 with exogenous cholesterol did not alter its infectivity (Fig. 1F, right) or change its density gradient (data not shown). These results suggest that exogenous cholesterol supplementation can reverse deficits in the infectivity of HCV virions due to low cholesterol content.
Sphingolipid dependence of HCV infectivity.
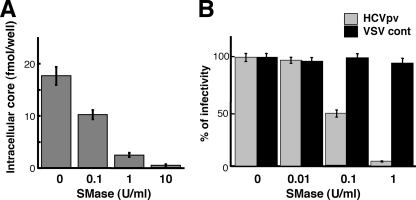
In addition to cholesterol, sphingolipid is a major component of eukaryotic lipid membranes. We therefore investigated the functional significance of sphingomyelin (SM), the most abundant sphingolipid, with regard to HCV infectivity. HCVcc was treated for 1 h with increasing concentrations (0.1 to 10 U/ml) of bacterial SMase, which is known to hydrolyze membrane-bound SM to ceramide. Following ultracentrifugation to remove the SMase, Huh-7 cells were inoculated with the HCVcc. The amount of HCV core protein within the cells was quantified at 72 h p.i. Figure 2A shows 50 and 90% reductions in HCV infectivity after incubation of the virion with 0.1 and 1 U/ml SMase, respectively. We further observed that SMase treatment of HCVpv resulted in a progressive loss of infectivity, while SMase had no effect on the infectivity of the control virus (Fig. 2B). This demonstrates that sphingolipid, like cholesterol, plays an essential role in HCV infectivity.
FIG. 2.
Effect of SM hydrolysis on viral infectivity. (A) Effect on the infectivity of HCVcc. HCVcc was treated with 0.1, 1, or 10 U/ml SMase for 1 h at 37°C, after which SMase was removed by ultracentifugation. Huh-7 cells were infected with the treated virus, and the core protein content of infected cells was determined at 72 h p.i. (B) Effect on the infectivity of HCVpv (genotype 1a) (shaded bars) or the control, VSVdelG-GFP/G (VSV cont) (solid bars). The viruses were preincubated with SMase for 1 h at 37°C before infection. Data are means from four independent experiments. Error bars, standard deviations.
Requirement for virion-associated cholesterol and sphingolipid during HCV cell entry.
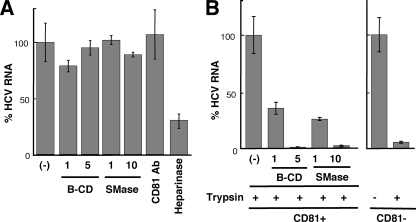
These findings support the idea that virion-associated cholesterol and sphingolipid may influence viral entry into host cells by altering the interaction between viral particles and a host cell factor(s). Viral entry is a multistep process including binding of the virion to the cell surface and internalization into the cytoplasm by endocytosis. To examine whether virion-associated cholesterol and SM might play a role in cell binding or postbinding events during viral entry, we used a binding assay in which Huh-7 cells preincubated for 1 h at 4°C were infected with B-CD- or SMase-treated HCVcc. Total RNA was extracted after a 1-h addition of the virions at 4°C, followed by quantification of HCV RNA. As shown in Fig. 3A, treatment of the virions with either B-CD or SMase had little influence on their ability to bind to cells.
FIG. 3.
Effects of B-CD or SMase on virus attachment and internalization. (A) Virus attachment to Huh-7 cells was determined at 4°C after treatment of HCVcc with B-CD (1 or 5 mg/ml) or SMase (1 or 10 U/ml). An antibody (Ab) against CD81 was used, in order to ensure that the antibody did not inhibit HCVcc binding (7, 33). Heparinase was used to reduce HCV attachment to the cell. Viral RNA copies were normalized to total cellular RNA, and the normalized RNA copies in the mock-treated sample (−) were arbitrarily set at 100%. (B) Virus internalization was measured in Huh7-25, a CD81-negative subclone (CD81−) (3), and Huh7-25-CD81, which stably expresses CD81 (CD81+), after treatment of the virions with B-CD or SMase. After internalization for 2 h at 37°C, cells were exposed to trypsin (trypsin +) or phosphate-buffered saline (trypsin −). Huh7-25 was used to ensure that surface-bound virus would be removed by trypsin treatment. The amounts of HCV RNA in Huh7-25 and Huh7-25-CD81 cells infected with untreated HCVcc were assigned the arbitrary value of 100%, respectively. Results are representative of four independent experiments.
It has been shown that CD81 plays an important role in HCV internalization but is not correlated with viral attachment (7, 33). An anti-CD81 antibody was used as a negative control for reduced viral attachment. It is likely that heparan sulfate proteoglycan on the target cell surface is needed for the initial attachment of HCV (33). Thus, heparinase I was used as a positive control for reduced HCV attachment to the cells. To examine the roles of cholesterol and sphingolipid on the HCVcc membrane in viral internalization, a virus-cell mixture prepared at 4°C as described above was incubated for 2 h at 37°C, followed by trypsinization to remove virions that were surface bound but not internalized (Fig. 3B). We verified that 94% of surface-bound-viruses were removed by trypsinization using CD81-negative Huh-7 subclones. A marked reduction in viral RNA levels within cells was detected after pretreatment of the virus with either B-CD or SMase. These results strongly suggest that virion-associated cholesterol and sphingolipid function as key determinants of internalization but not of cell attachment.
Association of HCV structural proteins with lipid rafts.
Cholesterol and sphingolipid are major components of lipid rafts, which can be isolated as detergent-resistant membranes (DRMs) by treatment with cold TX-100, followed by equilibrium flotation centrifugation. Matto et al. (30) reported that HCV core protein is associated with DRMs in cells carrying the full-length HCV replicon. To investigate whether HCV structural proteins are associated with DRMs in HCVcc-producing cells, lysates from cells infected with HCVcc were subjected to membrane flotation analysis. In the absence of detergent treatment, the majority of the core (Fig. 4A) and E1 (Fig. 4B) proteins were detected in the membrane fractions. After treatment with cold TX-100, significant amounts of both viral proteins were recovered from the DRM fraction. However, after treatment with TX-100 at 37°C, the majority of the E1 and core proteins had shifted to the detergent-soluble fractions. We also found that HCV genotype 1b E1 and E2 can be associated with the lipid raft in 293T cells transfected with an E1 or E2 expression plasmid (Fig. 4C) and that the cytoplasmic tails of envelope proteins are important for their interaction (Fig. 4D). These data suggest that subpopulations of HCV structural proteins are associated with lipid rafts in cells generating the HCV particles.
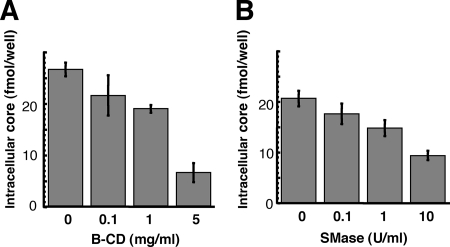
Moderate inhibition of HCV infection by B-CD or SMase treatment of host cells.
It has recently been reported that cholesterol depletion or SM hydrolysis from the host cell membrane decreases HCV infection, in part by decreasing the level of CD81 on the cell surface (19, 53). The involvement of the lipid environment of the host cell plasma membrane in HCV infection was investigated in our HCVcc infection system. Prior to infection, Huh-7 cells were treated with B-CD or SMase and then washed with the medium five times. Cholesterol depletion from Huh-7 cells by B-CD at 1 or 5 mg/ml inhibited HCV core levels by 20 and 75%, respectively, compared to levels in untreated cells (Fig. 5A). We also found that hydrolysis of SM by SMase at 1 or 10 U/ml on the cells, respectively, led to moderate reduction of the viral infection, by 20 or 55% of the infection level of the untreated control (Fig. 5B). There was no influence on cell viability under the conditions of these treatments (data not shown). These findings, compared with the results in Fig. 1A and 2A, suggest that the raft-like environment on the plasma membrane likely serves as a portal for HCV entry, but HCV virion-associated cholesterol and sphingolipid more readily play more critical roles in viral infection.
FIG. 5.
Effects of B-CD or SMase treatment of cells on HCV infectivity. Huh-7 cells were either left untreated or treated with B-CD at 0.1, 1, or 5 mg/ml (A) or with SMase at 0.1, 1, or 10 U/ml (B) prior to HCVcc infection. Intracellular core levels were quantitated 72 h p.i. Data are means from four independent experiments. Error bars, standard deviations.
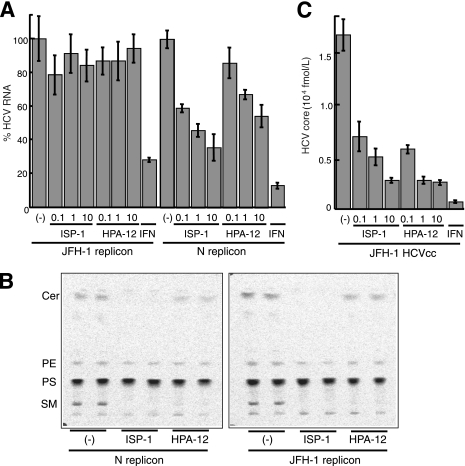
Inhibitors of the sphingolipid biosynthetic pathway suppress the production of HCVcc, but not RNA replication, for a JFH-1-derived replicon.
In the course of studying the involvement of lipid metabolism in the HCV life cycle, we observed that inhibitors of the sphingolipid biosynthetic pathway, including ISP-1 and HPA-12, which specifically inhibit serine palmitoyltransferase (31) and ceramide trafficking from the ER to the Golgi apparatus (55), influenced subgenomic replicons derived from the HCV-N isolate (genotype 1b), but not those derived from JFH-1. A dose-dependent decrease in HCV RNA copy numbers among HCV-N replicon cells was observed upon exposure to ISP-1 or HPA-12, as previously reported (43, 52). In contrast, these compounds had little or no effect on viral RNA accumulation in JFH-1 replicon cells (Fig. 6A). Furthermore, these compounds did not affect luciferase activity in the lysates of Huh-7 cells transfected with an in vitro-transcribed JFH-1 replicon RNA containing a luciferase reporter gene (22) (data not shown). Figure 6B shows the effects of ISP-1 and HPA-12 on de novo sphingolipid biosynthesis by replicon cells. No differences in the inhibitory effects of each compound were observed in replicon cells derived from HCV-N versus JFH-1. When de novo synthesis of sphingolipids was examined by metabolic labeling with [14C]serine, ISP-1 almost completely inhibited the production of both ceramide and SM, while HPA-12 greatly inhibited the synthesis of SM but not ceramide. Levels of phosphatidylethanolamine and phosphatidylserine, into which serine is incorporated by a pathway distinct from that of sphingolipid biosynthesis, were not influenced by these drugs. These results suggest that suppression of HCV RNA replication by inhibitors of sphingolipid biosynthesis might be dependent on the viral genotype or isolate.
FIG. 6.
Anti-HCV effects of inhibitors of the sphingolipid biosynthetic pathway. Subgenomic replicon cells derived from HCV isolate N or JFH-1, as well as HCVcc-producing cells, were treated with ISP-1 (0.1, 1, or 10 μM), HPA-12 (0.1, 1, or 10 μM) or alpha interferon (IFN) (100 U/ml) for 72 h. HCV RNA titers in the replicon cells (A) and the HCV core protein content of the culture medium of infected cells (C) were determined. Data are means from four independent experiments. Error bars, standard deviations. (B) De novo synthesis of sphingolipid in the absence or presence of ISP-1 (10 μM) and HPA-12 (10 μM) was monitored in duplicate by metabolic labeling with [14C]serine for 2 h at 37°C. Cer, ceramide; PE, phosphatidylethanolamine; PS, phosphatidylserine.
This observation prompted us to investigate whether inhibitors of the sphingolipid biosynthetic pathway might have the ability to prevent HCV virion production. Interestingly, when Huh-7 cells producing JFH-1 HCVcc were treated with ISP-1 or HPA-12 under conditions similar to those the replicon cells, viral core levels in the culture supernatants were greatly reduced in a dose-dependent manner. For example, exposure to 10 μM ISP-1 or 1 μM HPA-12 reduced viral core protein levels more than 85% from those for control cells (Fig. 6C). The 50% inhibitory concentrations of both drugs were less than 0.1 μM, 50-fold less than those obtained for the RNA replication of the HCV-N-replicon. Together, these results suggest that the sphingolipid biosynthetic pathway plays an important role in the production of HCV particles, but not in genome replication, in JFH-1-based HCVcc.
DISCUSSION
In this study, we demonstrated the role of HCV virion-associated cholesterol and sphingolipid in viral infectivity. Although dependence on virion-associated cholesterol for virus entry has been shown for a number of viruses (4, 6, 28, 49), this is the first study to demonstrate the importance of envelope cholesterol in a virus belonging to the family Flaviviridae. Furthermore, to our knowledge, the functional role of virion membrane-associated SM has not been examined in viruses. Our previous studies using Chinese hamster ovary cell mutants deficient in SM synthesis have demonstrated that reduction of cellular SM levels enhances cellular cholesterol efflux in the presence of B-CD (9, 12). Thus, it may be possible that SM plays a role in the retention of cholesterol on HCV particles due to interaction between cholesterol and SM. The finding that B-CD or SMase treatment of HCVcc markedly inhibited virus internalization but not cell attachment (Fig. 3) suggests that HCV membrane-associated cholesterol and sphingolipid are crucial for the interaction of viral glycoproteins with the virus-receptor/coreceptor required for cell entry. Cholesterol depletion or sphingolipid hydrolysis might induce a conformational change in the viral envelope, resulting in instability of the virion structure. Since the cholesterol/phospholipid ratios of membranes affect bilayer fluidity, the maturation of viral envelopes with high cholesterol/phospholipid ratios via association with rafts may be important for the stability of HCV particles. Replenishing the viral membrane with cholesterol following treatment with 5 mg/ml B-CD successfully restored viral infectivity to the same level as that of untreated virus (Fig. 1), suggesting that reversible B-CD-induced changes in HCV structure might critically influence viral infectivity. However, we were unable to restore viral infectivity by replenishing cholesterol after pretreatment of the virion with concentrations of B-CD exceeding 10 mg/ml (data not shown). Under these conditions, it is likely that large holes in the viral membrane destroy the virus, a result that cannot be reversed by supplying exogenous cholesterol.
How are cholesterol and sphingolipid involved in the HCV virion during the process of virus maturation? Like most positive-stranded RNA viruses, HCV is thought to assemble at the ER membrane. However, Miyanari et al. (32) reported that lipid droplets are important for HCVcc formation. These authors have shown that the characteristics of lipid-droplet-associated membranes in Huh-7 cells differ from those of ER membranes. In the case of flaviviruses, for which the mechanism of viral assembly and budding remains unclear (15), a few studies have demonstrated budding at the plasma membrane (13, 36, 37, 41), and it has been proposed that the site of budding may be virus and cell type dependent (27). We demonstrate here that subpopulations of HCV structural proteins partition into cellular detergent-resistant, lipid-raft-like membrane fractions in HCVcc-producing cells (Fig. 4) and that inhibitors of the sphingolipid biosynthetic pathway block HCV virion production (Fig. 6). Furthermore, a large proportion of HCV E2 protein incorporated into HCVcc is endoglycosidase H resistant (data not shown). Thus, membrane compartments containing cholesterol- and sphingolipid-rich microdomains may be involved in HCV virion maturation. Another explanation for the recruitment of these lipids to the HCV membrane may be an association between the virus and very-low-density lipoprotein (VLDL) or low-density lipoprotein. Recently, Huang et al. (16) demonstrated a close link between HCV production and VLDL assembly, suggesting that an HCV-VLDL complex is generated and secreted from cells.
Recent reports have demonstrated that CD81-mediated HCV infection is partly dependent on cell membrane cholesterol (19) and SM (53). We further characterized the role of lipid on the plasma membrane in viral infectivity and found that cholesterol depletion by B-CD, as well as hydrolysis of SM by SMase, moderately inhibits HCV infectivity (Fig. 5). These results suggest that cholesterol and sphingolipid in the plasma membrane environment may assist HCV entry, while HCV virion-associated cholesterol and sphingolipid appear to play critical roles in viral infection.
We previously demonstrated that HCV RNA and nonstructural proteins are present in DRM structures, likely in the context of a lipid-raft structure, and that viral RNA is likely synthesized at a raft membrane structure in cells containing the genotype 1b HCV replicon (2, 10, 46). Here we observed that ISP-1 and HPA-12 suppress HCVcc production, but not viral RNA replication, by the JFH-1 replicon (Fig. 6). Impairment of particle assembly and maturation, rather than suppression of genome replication, by these drugs may account for the inhibition of HCV production in the JFH-1 system. Viral RNA replication of the HCV-N replicon, however, was efficiently inhibited by these compounds, as found in previous reports (43). The virus strain specificity of the anti-HCV activity of cyclosporine has recently been demonstrated: JFH-1 replication is less sensitive to cyclosporine than replication of genotype 1b strains. Furthermore, the requirement for interaction with a cellular replication cofactor, cyclophilin B, differs among HCV strains (18). It appears that ISP-1 and HPA-12 are further examples of diverse effects on HCV strain replication.
In summary, our data here demonstrate important roles of cholesterol and sphingolipid in HCV infection and virion maturation. Specifically, mature HCV particles are rich in cholesterol. Depletion from HCV or hydrolysis of virion-associated SM results in a loss of infectivity. Moreover, the addition of exogenous cholesterol restores infectivity. In addition, cholesterol and sphingolipid on the HCV membrane play key roles in virus internalization, and portions of structural proteins are localized at lipid-raft-like membrane structures within cells. Finally, inhibitors of the sphingolipid biosynthetic pathway efficiently block virion production. These observations suggest that agents capable of modifying virion-associated lipid content might function as antivirals by preventing and/or blocking HCV infection and production.
Acknowledgments
We thank M. Matsuda, M. Sasaki, S. Yoshizaki, T. Shimoji, M. Kaga, and T. Date for technical assistance and T. Mizoguchi for secretarial work.
This work was partially supported by a grant-in-aid for Scientific Research from the Japan Society for the Promotion of Science, from the Ministry of Health, Labor, and Welfare of Japan, and from the Ministry of Education, Culture, Sports, Science, and Technology, as well as by a Research on Health Science Focusing on Drug Innovation grant from the Japan Health Sciences Foundation.
Footnotes
Published ahead of print on 26 March 2008.
REFERENCES
- 1.Aizaki, H., Y. Aoki, T. Harada, K. Ishii, T. Suzuki, S. Nagamori, G. Toda, Y. Matsuura, and T. Miyamura. 1998. Full-length complementary DNA of hepatitis C virus genome from an infectious blood sample. Hepatology 27621-627. [DOI] [PubMed] [Google Scholar]
- 2.Aizaki, H., K. J. Lee, V. M. Sung, H. Ishiko, and M. M. Lai. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324450-461. [DOI] [PubMed] [Google Scholar]
- 3.Akazawa, D., T. Date, K. Morikawa, A. Murayama, M. Miyamoto, M. Kaga, H. Barth, T. F. Baumert, J. Dubuisson, and T. Wakita. 2007. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J. Virol. 815036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, F. C., J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2005. Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry. J. Virol. 7911588-11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchard, E., D. Brand, S. Trassard, A. Goudeau, and P. Roingeard. 2002. Hepatitis C virus-like particle morphogenesis. J. Virol. 764073-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chazal, N., and D. Gerlier. 2003. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, M. J., T. von Hahn, D. M. Tscherne, A. J. Syder, M. Panis, B. Wolk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446801-805. [DOI] [PubMed] [Google Scholar]
- 8.Ezelle, H. J., D. Markovic, and G. N. Barber. 2002. Generation of hepatitis C virus-like particles by use of a recombinant vesicular stomatitis virus vector. J. Virol. 7612325-12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukasawa, M., M. Nishijima, H. Itabe, T. Takano, and K. Hanada. 2000. Reduction of sphingomyelin level without accumulation of ceramide in Chinese hamster ovary cells affects detergent-resistant membrane domains and enhances cellular cholesterol efflux to methyl-β-cyclodextrin. J. Biol. Chem. 27534028-34034. [DOI] [PubMed] [Google Scholar]
- 10.Gao, L., H. Aizaki, J. W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 783480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 758516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanada, K., T. Hara, M. Fukasawa, A. Yamaji, M. Umeda, and M. Nishijima. 1998. Mammalian cell mutants resistant to a sphingomyelin-directed cytolysin. Genetic and biochemical evidence for complex formation of the LCB1 protein with the LCB2 protein for serine palmitoyltransferase. J. Biol. Chem. 27333787-33794. [DOI] [PubMed] [Google Scholar]
- 13.Hase, T., P. L. Summers, K. H. Eckels, and W. B. Baze. 1987. An electron and immunoelectron microscopic study of dengue-2 virus infection of cultured mosquito cells: maturation events. Arch. Virol. 92273-291. [DOI] [PubMed] [Google Scholar]
- 14.Heider, J. G., and R. L. Boyett. 1978. The picomole determination of free and total cholesterol in cells in culture. J. Lipid Res. 19514-518. [PubMed] [Google Scholar]
- 15.Heinz, F. X., and S. L. Allison. 2003. Flavivirus structure and membrane fusion. Adv. Virus Res. 5963-97. [DOI] [PubMed] [Google Scholar]
- 16.Huang, H., F. Sun, D. M. Owen, W. Li, Y. Chen, M. Gale, and J. Ye. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. USA 1045848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 762997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishii, N., K. Watashi, T. Hishiki, K. Goto, D. Inoue, M. Hijikata, T. Wakita, N. Kato, and K. Shimotohno. 2006. Diverse effects of cyclosporine on hepatitis C virus strain replication. J. Virol. 804510-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapadia, S. B., H. Barth, T. Baumert, J. A. McKeating, and F. V. Chisari. 2007. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J. Virol. 81374-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, T., A. Furusaka, M. Miyamoto, T. Date, K. Yasui, J. Hiramoto, K. Nagayama, T. Tanaka, and T. Wakita. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J. Med. Virol. 64334-339. [DOI] [PubMed] [Google Scholar]
- 21.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 1251808-1817. [DOI] [PubMed] [Google Scholar]
- 22.Kato, T., T. Date, M. Miyamoto, M. Sugiyama, Y. Tanaka, E. Orito, T. Ohno, K. Sugihara, I. Hasegawa, K. Fujiwara, K. Ito, A. Ozasa, M. Mizokami, and T. Wakita. 2005. Detection of anti-hepatitis C virus effects of interferon and ribavirin by a sensitive replicon system. J. Clin. Microbiol. 435679-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, T., T. Date, A. Murayama, K. Morikawa, D. Akazawa, and T. Wakita. 2006. Cell culture and infection system for hepatitis C virus. Nat. Protoc. 12334-2339. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi. S, K. Kakumoto, and M. Sugiura. 2002. Transition metal salts-catalyzed aza-Michael reactions of enones with carbamates. Org. Lett. 181319-1322. [DOI] [PubMed] [Google Scholar]
- 25.Koutsoudakis, G., E. Herrmann, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 27.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 7510787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manes, S., G. del Real, R. A. Lacalle, P. Lucas, C. Gomez-Mouton, S. Sanchez-Palomino, R. Delgado, J. Alcami, E. Mira, and A. C. Martinez. 2000. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO 1190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuo, E., H. Tani, C. Lim, Y. Komoda, T. Okamoto, H. Miyamoto, K. Moriishi, S. Yagi, A. H. Patel, T. Miyamura, and Y. Matsuura. 2006. Characterization of HCV-like particles produced in a human hepatoma cell line by a recombinant baculovirus. Biochem. Biophys. Res. Commun. 340200-208. [DOI] [PubMed] [Google Scholar]
- 30.Matto, M., C. M. Rice, B. Aroeti, and J. S. Glenn. 2004. Hepatitis C virus core protein associates with detergent-resistant membranes distinct from classical plasma membrane rafts. J. Virol. 7812047-12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyake, Y., Y. Kozutsumi, S. Nakamura, T. Fujita, and T. Kawasaki. 1995. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem. Biophys. Res. Commun. 211396-403. [DOI] [PubMed] [Google Scholar]
- 32.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 91089-1097. [DOI] [PubMed] [Google Scholar]
- 33.Morikawa, K., Z. Zhao, T. Date, M. Miyamoto, A. Murayama, D. Akazawa, J. Tanabe, S. Sone, and T. Wakita. 2007. The roles of CD81 and glycosaminoglycans in the adsorption and uptake of infectious HCV particles. J. Med. Virol. 79714-723. [DOI] [PubMed] [Google Scholar]
- 34.Murakami, K., K. Ishii, Y. Ishihara, S. Yoshizaki, K. Tanaka, Y. Gotoh, H. Aizaki, M. Kohara, H. Yoshioka, Y. Mori, N. Manabe, I. Shoji, T. Sata, R. Bartenschlager, Y. Matsuura, T. Miyamura, and T. Suzuki. 2006. Production of infectious hepatitis C virus particles in three-dimensional cultures of the cell line carrying the genome-length dicistronic viral RNA of genotype 1b. Virology 351381-392. [DOI] [PubMed] [Google Scholar]
- 35.Nakai, K., T. Okamoto, T. Kimura-Someya, K. Ishii, C. K. Lim, H. Tani, E. Matsuo, T. Abe, Y. Mori, T. Suzuki, T. Miyamura, J. H. Nunberg, K. Moriishi, and Y. Matsuura. 2006. Oligomerization of hepatitis C virus core protein is crucial for interaction with the cytoplasmic domain of E1 envelope protein. J. Virol. 8011265-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng, M. L., J. Howe, V. Sreenivasan, and J. J. Mulders. 1994. Flavivirus West Nile (Sarafend) egress at the plasma membrane. Arch. Virol. 137303-313. [DOI] [PubMed] [Google Scholar]
- 37.Ng, M. L., S. H. Tan, and J. J. Chu. 2001. Transport and budding at two distinct sites of visible nucleocapsids of West Nile (Sarafend) virus. J. Med. Virol. 65758-764. [DOI] [PubMed] [Google Scholar]
- 38.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108193-199. [DOI] [PubMed] [Google Scholar]
- 39.Pessin, J. E., and M. Glaser. 1980. Budding of Rous sarcoma virus and vesicular stomatitis virus from localized lipid regions in the plasma membrane of chicken embryo fibroblasts. J. Biol. Chem. 2559044-9050. [PubMed] [Google Scholar]
- 40.Pitha, J., T. Irie, P. B. Sklar, and J. S. Nye. 1988. Drug solubilizers to aid pharmacologists: amorphous cyclodextrin derivatives. Life Sci. 43493-502. [DOI] [PubMed] [Google Scholar]
- 41.Rahman, S., T. Matsumura, K. Masuda, K. Kanemura, and T. Fukunaga. 1998. Maturation site of dengue type 2 virus in cultured mosquito C6/36 cells and Vero cells. Kobe J. Med. Sci. 4465-79. [PubMed] [Google Scholar]
- 42.Rouser, G., G. Galli, and G. Kritchevsky. 1967. Lipid composition of the normal human brain and its variations during various diseases. Pathol. Biol. 15195-200. [PubMed] [Google Scholar]
- 43.Sakamoto, H., K. Okamoto, M. Aoki, H. Kato, A. Katsume, A. Ohta, T. Tsukuda, N. Shimma, Y. Aoki, M. Arisawa, M. Kohara, and M. Sudoh. 2005. Host sphingolipid biosynthesis as a target for hepatitis C virus therapy. Nat. Chem. Biol. 1333-337. [DOI] [PubMed] [Google Scholar]
- 44.Sato, K., H. Okamoto, S. Aihara, Y. Hoshi, T. Tanaka, and S. Mishiro. 1993. Demonstration of sugar moiety on the surface of hepatitis C virions recovered from the circulation of infected humans. Virology 196354-357. [DOI] [PubMed] [Google Scholar]
- 45.Serafino, A., M. B. Valli, F. Andreola, A. Crema, G. Ravagnan, L. Bertolini, and G. Carloni. 2003. Suggested role of the Golgi apparatus and endoplasmic reticulum for crucial sites of hepatitis C virus replication in human lymphoblastoid cells infected in vitro. J. Med. Virol. 7031-41. [DOI] [PubMed] [Google Scholar]
- 46.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 774160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinitzky, M., and M. Inbar. 1976. Microviscosity parameters and protein mobility in biological membranes. Biochim. Biophys. Acta 433133-149. [DOI] [PubMed] [Google Scholar]
- 48.Shirakura, M., K. Murakami, T. Ichimura, R. Suzuki, T. Shimoji, K. Fukuda, K. Abe, S. Sato, M. Fukasawa, Y. Yamakawa, M. Nishijima, K. Moriishi, Y. Matsuura, T. Wakita, T. Suzuki, P. M. Howley, T. Miyamura, and I. Shoji. 2007. E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. J. Virol. 811174-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart, A. D., H. E. Eustace, T. A. McKee, and T. D. Brown. 2002. A novel cell entry pathway for a DAF-using human enterovirus is dependent on lipid rafts. J. Virol. 769307-9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takikawa, S., K. Ishii, H. Aizaki, T. Suzuki, H. Asakura, Y. Matsuura, and T. Miyamura. 2000. Cell fusion activity of hepatitis C virus envelope proteins. J. Virol. 745066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tani, H., Y. Komoda, E. Matsuo, K. Suzuki, I. Hamamoto, T. Yamashita, K. Moriishi, K. Fujiyama, T. Kanto, N. Hayashi, A. Owsianka, A. H. Patel, M. A. Whitt, and Y. Matsuura. 2007. Replication-competent recombinant vesicular stomatitis virus encoding hepatitis C virus envelope proteins. J. Virol. 818601-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Umehara, T., M. Sudoh, F. Yasui, C. Matsuda, Y. Hayashi, K. Chayama, and M. Kohara. 2006. Serine palmitoyltransferase inhibitor suppresses HCV replication in a mouse model. Biochem. Biophys. Res. Commun. 34667-73. [DOI] [PubMed] [Google Scholar]
- 53.Voisset, C., M. Lavie, F. Helle, A. Op De Beeck, A. Bilheu, J. Bertrand-Michel, F. Tercé, L. Cocquerel, C. Wychowski, N. Vu-Dac, and J. Dubuisson. 2008. Ceramide enrichment of the plasma membrane induces CD81 internalization and inhibits hepatitis C virus entry. Cell. Microbiol. 10606-617. [DOI] [PubMed] [Google Scholar]
- 54.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yasuda, S., H. Kitagawa, M. Ueno, H. Ishitani, M. Fukasawa, M. Nishijima, S. Kobayashi, and K. Hanada. 2001. A novel inhibitor of ceramide trafficking from the endoplasmic reticulum to the site of sphingomyelin synthesis. J. Biol. Chem. 27643994-44002. [DOI] [PubMed] [Google Scholar]
- 56.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 1029294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]