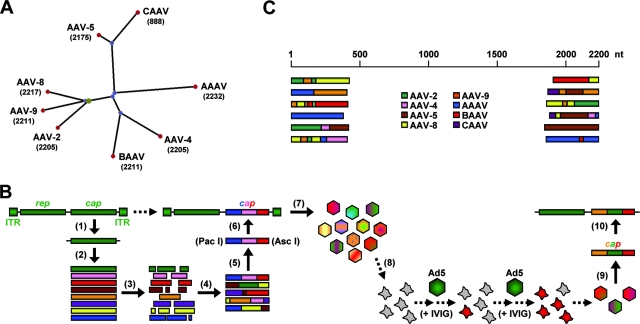
FIG. 1.
Generation of an AAV capsid library via DNA family shuffling. (A) Phylogram tree (created using PhyloDraw [http://pearl.cs.pusan.ac.kr/phylodraw/#test]) showing the eight AAV serotypes used as parents for DNA family shuffling (numbers denote lengths of capsid genes, in nucleotides). Branch lengths are proportional to the amounts of evolutionary change, calculated in ClustalW (http://www.ebi.ac.uk/clustalw/#). CAAV, AAAV, and BAAV, caprine, avian, and bovine AAVs, respectively. (B) Individual steps for generation of the library (scheme). Full-length cap genes were PCR amplified and subcloned for further amplification (1) and then isolated (2) and DNase I digested (3). Two consecutive PCRs without (4) or with (5) conserved primers were performed to reassemble shuffled full-length cap genes. These genes were inserted (6) into a plasmid carrying AAV-2 ITRs and a rep gene. The transfection of 293 cells with the resulting plasmid library (7) together with an adenoviral helper resulted in a viral library. One possible selection scheme used in this study was the coinfection of cultured liver cells (8) with the library and helper adenovirus under stringent conditions, resulting in the amplification of specific AAV capsids. Viral DNA can then be isolated (9) and cloned into an AAV helper plasmid carrying the AAV-2 rep gene without ITRs (10) for subsequent vector production. Ad5, helper adenovirus type 5. (C) Examples of shuffled cap genes in an initial small-scale library. DNA was extracted from 24 randomly chosen clones, and 5′ and 3′ ends of the individual cap genes were sequenced (using T3/T7 primers binding in the plasmid backbone). Shown, per end, are six representative alignments with the eight parents.