Abstract
The gene cluster composed of varicella-zoster virus (VZV) open reading frame 9 (ORF9) to ORF12 encodes four putative tegument proteins and is highly conserved in most alphaherpesviruses. In these experiments, the genes within this cluster were deleted from the VZV parent Oka (POKA) individually or in combination, and the consequences for VZV replication were evaluated with cultured cells in vitro and with human skin xenografts in SCID mice in vivo. As has been reported for ORF10, ORF11 and ORF12 were dispensable for VZV replication in melanoma and human embryonic fibroblast cells. In contrast, deletion of ORF9 was incompatible with the recovery of infectious virus. ORF9 localized to the virion tegument and formed complexes with glycoprotein E, which is an essential protein, in VZV-infected cells. Recombinants lacking ORF10 and ORF11 (POKAΔ10/11), ORF11 and ORF12 (POKAΔ11/12), or ORF10, ORF11 and ORF12 (POKAΔ10/11/12) were viable in cultured cells. Their growth kinetics did not differ from those of POKA, and nucleocapsid formation and virion assembly were not disrupted. In addition, these deletion mutants showed no differences compared to POKA in infectivity levels for primary human tonsil T cells. Deletion of ORF12 had no effect on skin infection, whereas replication of POKAΔ11, POKAΔ10/11, and POKAΔ11/12 was severely reduced, and no virus was recovered from skin xenografts inoculated with POKAΔ10/11/12. These results indicate that with the exception of ORF9, the individual genes within the ORF9-to-ORF12 gene cluster are dispensable and can be deleted simultaneously without any apparent effect on VZV replication in vitro but that the ORF10-to-ORF12 cluster is essential for VZV virulence in skin in vivo.
Varicella-zoster virus (VZV) is a human alphaherpesvirus that causes varicella, or chickenpox, as the primary infection, establishes latency in sensory ganglia, and may reactivate to result in herpes zoster (6). Mature VZV virions, like those of other herpesviruses, are composed of the nucleocapsid containing the double-stranded DNA genome, the tegument, and the envelope. In the alphaherpesviruses, the tegument appears to be made up of more than 15 virus-encoded proteins (28, 38). In VZV, the immediate-early 62 protein (IE62), which is the major viral transactivator, and other regulatory proteins, including the IE4, IE63, open reading frame 10 (ORF10), and viral kinase ORF47 proteins, have been experimentally determined to be tegument proteins, but the functions of many of the putative VZV teguments have not yet been defined (22, 23, 37).
A gene cluster corresponding to VZV ORF9, ORF10, ORF11, and ORF12 is located near the beginning of the unique long region of the VZV genome and is highly conserved in most alphaherpesviruses (9). Based on homologies with herpes simplex virus type 1 (HSV-1), which is the prototype alphaherpesvirus, the proteins encoded by the gene cluster are believed to be tegument proteins, although only ORF10 has been demonstrated to be a component of VZV virions (8). It is known that ORF10 also acts as a transactivator for enhancing IE62 expression (30), and ORF10 is dispensable for VZV replication in cultured cells when it is removed by mutagenesis of vaccine OKA or parent OKA (POKA) cosmids (3, 5). However, we demonstrated that ORF10 is required for normal VZV virion formation and virulence in human skin xenografts in the SCID-hu mouse model (3). Recent studies have shown that ORF9 protein interacts with IE62, suggesting that ORF9 may have a role in the recruitment of IE62 and possibly other tegument proteins for incorporation into virion tegument (4). In addition, disruption of the ORF9 start codon, by using an infectious bacterial artificial chromosome (BAC) clone of VZV POKA, was not compatible with VZV replication (39). Other than these reports, little is known about contributions of the ORF9-to-ORF12 gene cluster during VZV replication.
When the genes that correspond to the ORF9-to-ORF12 cluster in other alphaherpesviruses have been investigated for their functions in viral replication or virion assembly, their characteristics appear to be different depending upon the virus. For example, HSV-1 UL46 (VP11/12), the homolog of ORF12, UL47 (VP13/14), the homolog of ORF11, and UL49 (VP22), the homolog of ORF9, have been reported to be dispensable, but UL48 (VP16), the ORF10 homolog, is essential for virus replication (13, 14, 34, 41, 42, 43). In another example, removal of the complete UL46-UL49 gene cluster from the pseudorabies virus (PRV) genome is compatible with viral replication (16). Thus, while the gene cluster is conserved, there is evidence of functional divergence among the alphaherpesvirus subfamily. Therefore, the purpose of these experiments was to use cosmid mutagenesis to analyze the functions of this conserved gene cluster during VZV infection, expanding upon previous studies of ORF9 and ORF10 to include ORF11 and ORF12 and examining the potential redundancies of these genes by removing each individually and in combination from the VZV genome.
MATERIALS AND METHODS
Generation of cluster gene mutants.
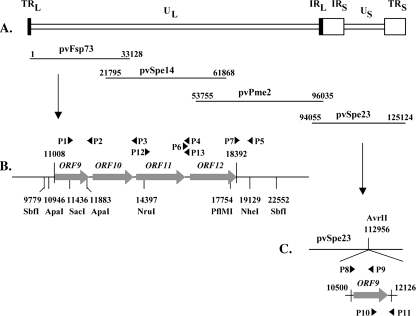
Four overlapping POKA cosmids, pvFsp73, pvSpe14, pvPme2, and pvSpe23, were used to make cluster gene deletions (Fig. 1A). The pvFsp73 cosmid was digested with ApaI enzyme and religated to remove the VZV ORF9 gene, which is located at nucleotides (nt) 11008 to 11916 of the genome (Fig. 1B). To delete ORF11, encoded by nt 13589 to 15970, the fragment from nt 11436 to 13587 was amplified by PCR with the forward primer P1 (5′-GGATTCCCGAAGCGAGCTC-3′), including a SacI restriction site, and the reverse primer P3 (5′-CGCATGAATTCTTATAACCGAGACAAACAAACG-3′), introducing an EcoRI restriction site; another fragment from nt 15988 to 19129 was amplified by PCR with the forward primer P6, containing an introduced EcoRI restriction site (5′-CGCATGAATTCATTAGGATGAAAAGAACGTTTCC-3′), and the reverse primer P5 (5′-GGGATCGAGCTAGCTCGAC-3′), including a NheI restriction site (underlined sequences show the positions of the restriction sites used for cloning). The resulting PCR products were digested with SacI and EcoRI and with EcoRI and NheI, respectively, and then were inserted by triple ligation into cosmid pvFsp73, which had been digested with SacI and NheI. Similar strategies were used to delete ORF10 and -11, ORF11 and -12, and ORF10 to -12 from pvFsp73. Two pairs of primers, P1 and P2 (5′-CGCATGGAATTCACTCCATACTTATA GAGTAAAATC-3′) and P5 and P6, were used to delete ORF10 and -11; one set of primers, P1 and P3 plus P5 and P7 (5′-CGCATGGAATTCGCGGTTAATAGATAG TTATGGAC-3′), were used to delete ORF11 and -12 (underlined sequences show the positions of the introduced EcoRI restriction sites). To remove ORF10 to -12 from pvFsp73, primers P1 and P2 and primers P5 and P7 were used. The resulting PCR products were digested by SacI and EcoRI or by EcoRI and NheI. Digested PCR fragments, as described above, were ligated back to cosmid pvFsp73, which was cut by SacI and NheI.
FIG. 1.
Mutagenesis of the VZV ORF9-to-ORF12 gene cluster. (A) Schematic diagram of the VZV genome and the overlapping cosmids pvFsp73, pvSpe14, pvPme2, and pvSpe23. The unique long (UL) and short (US) regions, the inverted sequences (internal repeat [IR] and terminal repeat [TR]) are indicated. (B) The genomic region of the cluster gene, including ORF9, -10, -11, and -12. The restriction enzyme sites used to generate deletions from the pvFsp73 cosmid are shown along with the locations of primers and their polarity (P1 to P7, P12, and P13; arrows). (C) Construction of the ORF9 rescue recombinant (POKA-ORF9-res). A fragment covering the genomic region from nt 10500 to nt 12126, which contains the complete ORF9 gene and its putative promoter and poly(A) site, was synthesized by PCR with primers P8 to P11 (arrows) and cloned into the unique AvrII site located at nt 112956 in the pvSpe23 cosmid.
To delete ORF12, which is located at nt 16135 to 18120 of the genome, a SbfI fragment containing nt 9779 to 22552 was cloned into a pET-Blue vector (Novagen, Madison, WI). The resulting vector is designated pETSbfI. By using pETSbfI as a template, PCR was performed with the forward primer P12 (5′-GCGCATAATGGGTTTGATGA-3′), corresponding to nt 14345 to 14365 of the VZV genome, and the reverse primer P13 (5′-TATAATTTGTCCATATCGCAtagCCACTGTTTGGACACTG-5′), including 20 nt of genome sequence, from nt 16115 to 16134, and a introduced stop codon (lowercase) and a PflMI restriction site (underlined). The resulting PCR product, which included nt 14245 to 16134, plus an introduced stop codon and a PflMI restriction site, was then digested by NruI and PfIMI. The NruI restriction site is located at nt 14397 of the VZV genome. The vector pETSbfI was also digested with NruI and PflMI restriction enzymes to release the fragment containing nt 14397 to 17754. The digested PCR fragment and the vector pETSbfI were ligated together to generate a plasmid containing a SbfI fragment with an introduced stop codon at the beginning of ORF12 and a deletion of 1,620 bp of ORF12. This vector is designated pETSbfIΔORF12. The SbfI fragment obtained by digesting the vector pETSbfIΔORF12 was ligated back to the cosmid pvFsp73, which was cut by SbfI. All PCR primers were synthesized either by Elim Biopharmaceuticals, Inc. (Hayward, CA), or by Biotech Core, Inc. (Mountain View, CA). The cosmid mutations were verified by sequencing.
Construction of the ORF9 rescue virus.
In order to restore ORF9, a fragment containing the intact ORF9 gene, the ORF9 putative promoter, and the poly(A) site was amplified and inserted to a unique AvrII site located in the pvSpe23 cosmid. This process was done using a three-step PCR because the ORF9 sequence has an AvrII site. The first PCR was done with primer P8 (5′-GTACTCGCCTAGGGTCGACTTACTTTATCGTCTAAC-3′), containing an introduced AvrII site, and antisense primer P9 (5′-CGATGCTCCTTCACCgAGGTCTGCTTCATTAGC-3′), including the targeted nucleotide “T” changed to the nucleotide “C” from ORF9 sequence (shown in lowercase italics for antisense primer P9); the second PCR used sense primer P10 (5′-GCTAATGAAGCAGACCTcGGTGAAGGAGCATCG-3′), which is complementary to primer P10, and antisense primer P11 (5′-GACGATGCCTAGGCGATTCCCTTTATCAAAACCCG-3′), containing an introduced AvrII site (underlined sequences show the positions of the introduced AvrII restriction sites). The resulting PCR products, which are complementary at their junction sites, were taken as templates for the third PCR, done with primers P8 and P11, to generate an AvrII-AvrII ORF9 fragment, in which the nucleotide “T” was switched to “C,” which did not change the ORF9 amino acid sequence. The AvrII-AvrII ORF9 fragment was cloned into a unique AvrII site of the pvSpe23 cosmid. Insertion was verified by sequencing (Elim Biopharmaceuticals, Inc., Hayward, CA).
Cells and recombinant viruses.
Recombinant viruses were isolated after transfection of human melanoma (MeWo) cells (21) with the mutated pvFsp73 cosmid and three intact cosmids, pvSpe14, pvPme2, and pvSpe23 (32). MeWo cells were transfected with three intact cosmids, pvFsp73, pvSpe14, pvPme2, together with the modified pvSpe23 containing ORF9 to generate the ORF9 rescue virus (POKA-ORF9-res). Melanoma cells were maintained in tissue culture medium (Eagle minimum essential medium; Mediatech, Washington, DC) supplemented with 10% fetal calf serum (Gemini Bio-Products, Woodland, CA), nonessential amino acids, and antibiotics. To confirm the deletions, DNA was recovered from infected cells or xenograft tissues with DNAzol reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. PCR was performed using Pfu DNA polymerase (Stratagene, La Jolla, CA) with different sets of primers flanking the regions of deletion or insertion. The resulting PCR products were electrophoresed on a 1% agarose gel, isolated with a QIAquick gel extraction kit (Qiagen, Inc., Valencia, CA), and confirmed by sequencing. Viruses were propagated in human embryonic lung fibroblast (HELF) cells and stored at −80° in fetal calf serum with 10% dimethyl sulfoxide for infection of SCID-hu mouse human skin xenografts.
The replication kinetics and peak titers of individual or combined cluster gene mutant viruses and of POKA-ORF9-res virus were assessed by infectious focus assays (29). Melanoma cells were seeded in six-well plates and infected with an inoculum of ∼1 × 103 PFU. On days 1 to 6, cells were trypsinized, centrifuged, and resuspended in 1 ml of culture medium. The infected cells were serially diluted 10-fold, and 0.1 ml of the solution was added to a melanoma monolayer in 24-well plates in triplicate. Cells were fixed in crystal violet in 20% ethanol, and plaques were counted with an inverted light microscope (magnification, ×4), Statistical differences in growth kinetics were determined by Student's t test.
Northern blots.
Northern blot procedures were performed as previously described (3). Briefly, two T75 (75-cm2) flasks of HELF cells were simultaneously infected with POKA and POK-ORF9-res, infected cells were harvested at a cytopathic effect of about 3, and total single-stranded RNAs were extracted with TRIzol reagent (Invitrogen, Carlsbad, CA) to perform Northern blotting. The preparations of riboprobes of ORF9A, ORF9, and ORF10 were described previously (3). The riboprobe for ORF8 was generated by primers P14 (5′-CAGTCTCTTATAATCGGTAAGAC-3′) and P15 (5′-GAACGAAGCGGTAA TTGATCC-3′), covering the region of the VZV genome from nt 9870 to 10664, and was synthesized by PCR using the pvFsp73 cosmid DNA as template. The specific PCR product was inserted into a pCR4-TOPO clone vector (Invitrogen, Carlsbad, CA) and was used to synthesize positive-stranded-RNA-specific probes by using Sp6-RNA polymerase (Ambion, Inc.).
Western blots and coimmunoprecipitation.
Whole-cell lysates were prepared by lysing POKA-infected or uninfected melanoma cells with 2 ml radioimmunoprecipitation assay buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% NP-40 [Sigma, St. Louis, MO], 0.1% sodium dodecyl sulfate [Bio-Rad, Hercules, CA], 1% sodium deoxycholic acid Σ, and a cocktail of protease inhibitor [Roche]) per T75 flask, followed by sonication and centrifugation.
Coimmunoprecipitation kits ExactaCruzE or ExactaCruzF (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) were used to investigate the possible complex formation between ORF9 and glycoprotein E (gE) in infected cells. Experiments were performed according to the manufacturer's protocols. Briefly, infected cell lysates were precleared by incubating with preclearing matrix for at least 1 h at 4°C while rotating. The preclearing matrix (beads) was pelleted by centrifugation, washed five times with phosphate-buffered saline (PBS), and resuspended in electrophoresis buffer (50 μl), while the supernatant of precleared cell lysates was used for immunoprecipitation. To form the immunoprecipitation antibody-bead complex, beads (50 μl) were conjugated with either anti-gE monoclonal antibody (MAb) 3B3 (36) or anti-ORF9 polyclonal antibody (kindly provided by W. T. Ruyechan, University of New York at Buffalo, Buffalo, NY) for 4 h at 4°C. The antibody-bound beads were washed with PBS and incubated with precleared lysates overnight at 4°C. Beads were washed and resuspended in electrophoresis buffer (50 μl).
The cell lysates, preclearing beads, and immunoprecipitation sample were boiled in electrophoresis buffer and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 7.5% gels. Proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore, Bedford, MA). Immunoblotting was performed using rabbit polyclonal antibody to ORF9 or mouse MAb to gE.
Infection of human tonsil cells.
Tonsils were kindly provided by the Department of Pathology, Stanford University Medical Center. Primary tonsil T cells were prepared from human tonsils and obtained as previously described (25). A monolayer of HELF cells in a T75 flask was infected with POKA or the deletion mutants and was overlaid with 5 × 106 purified T cells. At 48 h postinfection, T cells were collected for fluorescence-activated cell sorting (FACS) analysis, and the infected HELF cells were titered on melanoma cells. Uninfected HELF cells overlaid with purified T cells served as a negative control. The T cells (∼0.5 × 106 cells) were resuspended in 100 μl of FACS staining buffer (1% fetal bovine serum in 1× PBS). The T cells were then incubated with human immune or nonimmune polyclonal immunoglobulin G (IgG) on ice for 40 min, washed twice with FACS staining buffer, and stained with goat anti-human fluorescence isothiocyanate-conjugated (Caltag, Inc.) and mouse anti-human-CD3-phycoerythrin (Caltag, Inc.) antibodies for 40 min on ice, washed twice, and then fixed by 1% paraformaldehyde. The stained cells were analyzed with a FACSCalibur instrument (Becton Dickinson, Inc.).
Infection of human xenografts in SCID-hu mice.
Skin xenografts were engrafted in male homozygous C.B-17 scid mice (29) by using human fetal tissues obtained from Advanced Bioscience Resources (Alameda, CA) with informed consent according to federal and state regulations. Animals were cared for according to guidelines of the Animal Welfare Act, Public Law 94-279, and the Stanford University Administrative Panel on Laboratory Animal Care. VZV recombinant viruses propagated in HELF cells were used to inoculate xenografts. Infectious virus titers were determined for each inoculum at the time of injection. Skin xenografts were harvested after 10 and 21 days postinfection, homogenized, and resuspended in 1 ml of 1× PBS for infectious focus assays. Tissue suspensions were serially diluted 10-fold, and 0.1 ml of the solution was added to melanoma cells in 24-well plates in triplicate. Cells were fixed in 4% paraformaldehyde, incubated with polyclonal anti-VZV human immune serum and secondary anti-human biotin (Vector Laboratories, Inc., Burlingame, CA), and then stained with Fast Red substrate (Sigma). Statistical differences in growth kinetics were determined by Student's t test.
Immunohistochemistry.
Formalin-fixed, paraffin-embedded skin sections (5 μm) were deparaffinized and rehydrated. VZV protein was detected with human polyclonal anti-VZV IgG, biotinylated goat anti-human secondary antibody, and horseradish peroxidase-conjugated streptavidin (Chemicon IHC Select). Signals were developed with Vector VIP (purple) and counterstained with methyl green (Vector Laboratories, Inc., Burlingame, CA).
TEM.
HELF cells infected with POKA or the deletion mutants were trypsinized at 72 h postinfection, centrifuged, and immediately fixed with 2% glutaraldehyde in 0.1 M phosphate buffer (PBS), pH 7.0, for 2 h. The specimens were washed twice in PBS and postfixed with 1% osmium tetroxide (Polysciences, Inc., Warrington, PA) in PBS for 1 h, and after two 10-min washes in double distilled water, specimens were treated with 0.25% uranyl acetate (Polysciences, Inc.) overnight. After 24 h, the specimens were washed with water and dehydrated through a graded series of alcohol and propylene oxide washes. Each sample was infiltrated sequentially with 2:1 and 1:1 propylene oxide-Epon (Resolution Performance Products, Houston, TX) for 4 h, incubated overnight with 100% Epon, transferred to fresh Epon, and embedded and polymerized at 60°C for 24 h. Thin sections were collected on copper grids, stained with uranyl acetate and lead citrate, and viewed using a Phillips CM-12 transmission electron microscope (TEM).
In immunogold EM experiments, samples were prepared for cryosectioning or were cryofixed by high-pressure freezing. For cryosectioning, the cells were fixed in 4% paraformaldehyde with 0.1% glutaraldehyde in phosphate buffer (0.1 M; pH 7.2), washed several times in PBS, and infiltrated in 2.3 M sucrose overnight at 4°C. Samples were then mounted on pins for cryo-ultramicrotomy and frozen in liquid nitrogen. Ultrathin cryosections (80 nm) were prepared with a diamond knife (Diatome) at −130°C using an ultramicrotome (Ultracut; Leica) equipped with a cryosectioning chamber. Thawed cryosections were transferred to Formvar- and carbon-coated EM grids (Nickel) with a drop of 2.3 M sucrose and were immunolabeled (see below) and counterstained for EM with 0.5% uranylacetate in 2% methylcellulose for 10 min on ice. Samples, which were first high-pressure frozen with an EM-Pact2 (Leica, Vienna, Austria), were freeze substituted with 0.2% glutaraldehyde and 0.1% uranyl acetate in acetone and subsequently embedded in London Resin White (LR White). Ultrathin (70-nm) sections were then prepared as described for the standard EM procedure.
For immunogold labeling, the thawed cryosections and LR White sections were blocked with 1% fish gelatin in PBS for 30 min and incubated with a rabbit antibody to ORF9 for 1 h at room temperature, followed by incubation with protein A gold conjugates (15-nm gold particles) for 30 min. Sections were then counterstained as described above. Immunogold-labeled sections were visualized by a Jeol 1230 TEM at 80 kV, and digital photographs were taken with a Gatan Multiscan 701 digital camera.
RESULTS
Generation of ORF9-to-ORF12 cluster gene mutants.
To evaluate the contributions of the ORF9-to-ORF12 gene cluster to VZV replication, ORF9, ORF11, and ORF12 genes were deleted individually from the pvFsp73 cosmid (Fig. 1). Our POKAΔORF10 deletion mutant has been described previously (3). ORF9 has been identified as an essential gene by introducing a point mutation into the POKA BAC at the start codon of ORF9 (39). In these experiments, we removed the complete ORF9 coding sequence by cosmid mutagenesis. When each of the pvFsp73 cosmids from which ORF9, ORF11, or ORF12 had been deleted were transfected together with the other three intact POKA cosmids into melanoma cells, infectious virus was recovered from transfections with pvFsp73ΔORF11 and pvFsp73ΔORF12 but not with pvFsp73ΔORF9. Transfections done with two independently derived pvFsp73ΔORF9 cosmids were repeated three times with the same negative result. As a positive control, intact cosmids pvFsp73, pvSpe14, pvPme2, and pvSpe23 were cotransfected in parallel experiments and yielded infectious virus consistently, with plaques visible by 5 to 10 days after transfection. The viruses recovered from transfections of pvFsp73ΔORF11 and pvFsp73ΔORF12 were sequenced to demonstrate the expected mutations and were designated POKAΔ11 and POKAΔ12, respectively.
To examine whether the nonessential ORF10-to-ORF12 genes in the cluster might have interdependent or redundant functions, we generated pvFsp73 cosmids that had double or triple deletions of ORF10, -11 and -12. Two pvFsp73 cosmids from which either ORF10 and -11 or ORF11 and -12 had been removed were transfected into melanoma cells along with the other three intact cosmids. Both transfections yielded infectious viruses, which were designated POKAΔ10/11 and POKAΔ11/12 (Fig. 1B). Cotransfection of the pvFsp73 cosmid from which ORF10, ORF11, and ORF12 had been deleted and three intact cosmids also yielded infectious virus, designated POKAΔ10/11/12 (Fig. 1B).
ORF9 is essential for VZV replication.
To verify that the failure to recover infectious virus from transfections with pvFsp73ΔORF9 was caused by deleting ORF9, the ORF9 gene, along with its putative promoter and poly(A) site, was cloned into the unique AvrII site in the pvSpe23 cosmid (Fig. 1C). Cotransfection of this cosmid with the two intact cosmids, pvSpe14 and pvPme2, and pvFsp73ΔORF9 into melanoma cells yielded infectious virus, which was designated POKA-ORF9-res. The deletion of ORF9 from its native location and the insertion of ORF9 and its promoter at the nonnative site were confirmed by sequencing the recombinant virus.
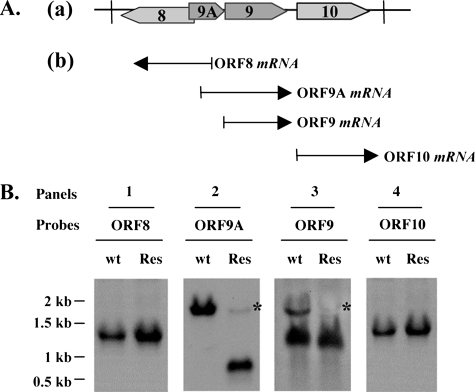
This POKA-ORF9-res virus was further characterized by investigating whether the expected ORF9 transcript was expressed from the nonnative site by Northern blot analysis (Fig. 2). In addition, to eliminate the concern that failure to recover infectious virus when ORF9 was deleted might have been due to effects on expression of its adjacent genes, the transcripts expressed from ORF9-adjacent genes were also analyzed by Northern blotting. Equal samples of each mRNA preparation, as judged with an ethidium bromide-stained ribosome RNA loading control (data not shown) were loaded in these Northern blots. According to a previous report, transcripts of ORF9 and ORF9A are 3′ coterminal, and their coding regions do not overlap (7, 35); thus, the termination signal of ORF9A was predicted to remain intact despite removal of the ORF9 coding region (Fig. 2A). The ORF9 probe detected two bands in total RNAs extracted from cells infected with POKA, one corresponding to ORF9 mRNA and the other to ORF9A mRNA, reading though ORF9A and ORF9. In contrast, when cells were infected with POKA-ORF9-res, a single strong band representing ORF9 mRNA was detected, indicating that ORF9 was expressed from the nonnative AvrII site in the rescue virus (Fig. 2B, panel 3). The probe covering ORF9A revealed a strong signal in cells infected with POKA, whereas a smaller transcript, of a size corresponding to that of the ORF9A coding region, was observed for cells infected with POKA-ORF9-res, suggesting that the removal of ORF9 from the VZV genome did not affect ORF9A expression (Fig. 2B, panel 2). Since the fragment inserted to rescue ORF9 included nt 10500 to 12126 of the VZV genome, which may contain a portion of the ORF9A promoter, a very faint signal was detected for mRNAs from cells infected with POKA-ORF9-res by using both ORF9 and ORF9A probes (Fig. 2B, panels 2 and 3). Experiments with probes for ORF8 and ORF10 demonstrated that deletion of ORF9 did not disrupt transcription from either the ORF8 or the ORF10 promoter, since the same gene expression patterns were observed for Northern blots with mRNAs from cells infected with POKA or POKA-ORF9-res (Fig. 2B, panels 1 and 4). In addition, the growth kinetics and plaque morphologies of POKA-ORF9-res and POKA were also indistinguishable (data not shown). Taken together, these experiments indicate that the failure to generate infectious virus was due specifically to the deletion of ORF9 and confirm that ORF9 is required for VZV replication.
FIG. 2.
Transcription of ORF9 and adjacent genes in cells infected with POKA and POKA-ORF9-res viruses. (A) Diagram of the genomic organization of VZV ORF8 to ORF10 and the direction of their transcription (a). Transcripts corresponding to these ORFs are represented by arrows below the diagram of the VZV ORF8-to-ORF10 segment (b). (B) Northern blots were done using riboprobes specific to ORF8 (panel 1), ORF9A (panel 2), ORF9 (panel 3), and ORF10 (panel 4), respectively, to examine the expression of these genes in HELF cells infected with POKA (wild-type [wt]) and POKA-ORF9-res (Res) viruses. The very faint ORF9A mRNA signal (indicated by asterisks) detected by both ORF9 and ORF9A probes in POKA-ORF9-res was generated from the insertion of the ORF9 rescue fragment and is indicated by an asterisk (panels 2 and 3). The numbers to the left are RNA kilobase size markers.
ORF9 is a component of VZV virions and binds to gE in VZV-infected cells.
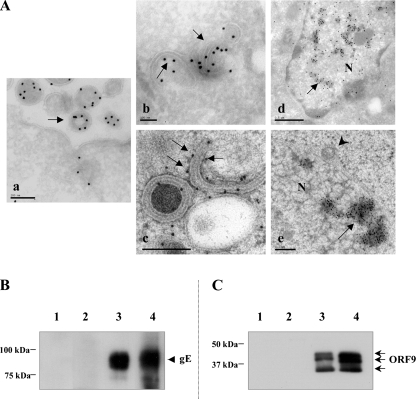
Cells infected with POKA were fixed, cryosectioned, incubated with a rabbit anti-ORF9 antibody followed by a secondary gold-linked antibody, and examined by TEM). ORF9 was detected in the tegument of virions (Fig. 3A, panel a). In addition, ORF9 was localized by immunogold EM to putative Golgi cisternae and Golgi-derived membranes (Fig. 3A, panel b). This observation was confirmed using another EM approach, in which the samples were high-pressure frozen and freeze substituted to preserve better cell ultrastructure and to visualize the locations of viral proteins; ORF9 was also associated with Golgi-like membranes under these conditions (Fig. 3A, panel c). Moreover, both EM approaches revealed that ORF9 appeared to accumulate together with electron-dense materials in the nuclei (Fig. 3A, panels d and e). ORF9 was not detected in association with primary enveloped virions located within the perinuclear space (Fig. 3A, panel e).
FIG. 3.
Intracellular localization of ORF9 and ORF9 interactions with gE in vitro. (A) Cells infected with POKA were fixed, embedded, cryosectioned, and then incubated with a rabbit antibody to VZV ORF9 followed by incubation with protein A gold conjugates (panels a, b, and d). ORF9 (15-nm gold particles) is incorporated into VZV virions and present in the virion tegument (a, arrow). ORF9 was also detected in association with putative Golgi cisternae and Golgi-derived membranes in the cytoplasm (b, arrows) and with electron-dense materials in the nucleus (d, arrow). When cells were prepared by high-pressure freezing, ORF9 was also detected in association with putative Golgi cisternae and Golgi-derived membranes in the cytoplasm (c, arrows) and with electron-dense materials in the nucleus (e, arrows). ORF9 is absent from perinuclear VZV virions that have a primary envelope (e, arrowhead). Bars, 0.2 μm (a and e), 0.1 μm (b and c), and 0.5 μm (d). N, nucleus. (B) Coimmunoprecipitation of the gE protein with anti-ORF9 rabbit polyclonal antibody. Melanoma cells were infected with VZV and harvested in 1% NP-40 lysis buffer, and cells extracts were incubated with beads coated with ORF9 antibody and Western blotted with anti-gE mouse MAb 3B3. Lane 1, uninfected cell extract; lane 2, beads alone; lane 3, infected cell extract; lane 4, beads coated with ORF9 antibody. (C) Coimmunoprecipitation of the ORF9 protein with beads coated with anti-gE MAb 3B3. Infected cells extracts were incubated with anti-gE MAb 3B3, and the membrane was probed with anti-ORF9 antibody. Lane 1, uninfected cell extract; lane 2, beads alone; lane 3, beads coated with gE antibody; lane 4, infected cell extract. Molecular mass markers are indicated on the left.
In order to investigate whether ORF9, like its HSV-1 homolog VP22, interacted with gE (15), coimmunoprecipitation was performed using VZV-infected melanoma cell extracts. gE was coprecipitated with ORF9 using anti-ORF9 polyclonal rabbit antibody (Fig. 3B), and ORF9 was coprecipitated with gE using anti-gE monoclonal mouse antibody (Fig. 3C). In a Western blot analysis of infected cell extracts and the sample coimmunoprecipitated by anti-gE antibody, ORF9 antibody specifically reacted with three polypeptides exhibiting apparent masses of about 33 kDa, 37 kDa, and 39 kDa (Fig. 3C, lanes 3 and 4), which likely correspond to the different forms of ORF9. These proteins were not detected in uninfected cell lysate or in the coimmunoprecipitation negative control (Fig. 3C, lanes 1 and 2). This experiment was repeated two more times, and the results were reproducible. These results indicated that gE interacts with ORF9 in VZV-infected cells, either directly or as part of a complex with other proteins.
Replication of the cluster gene deletion mutants in melanoma cells.
In these experiments, monolayers of melanoma cells were infected with approximately 1,000 PFU of POKA or the cluster gene deletion mutants to assess growth kinetics. The growth of POKA and that of the single-gene deletion mutants, POKAΔ11 and POKAΔ12, were similar (data not shown). The POKAΔ11 titer was 7.5 × 103 PFU/ml at day 1, which was slightly lower than those for POKA and POKAΔ12, but all titers were equivalent at 2 to 5 days after infection based on statistical analysis. The growth kinetics of POKAΔ10/11, POKAΔ11/12, and POKAΔ10/11/12 were also similar to those of POKA (data not shown). Although there was a minor reduction of POKAΔ10/11/12 at day 3 and 4 postinfection compared to POKA, the differences were not statistically significant (P > 0.05). In addition, the morphologies of plaques produced by the cluster gene mutants were indistinguishable from those produced by POKA. Plaque morphologies were also not different from those of POKA when the cluster gene mutants were grown in HELF cells.
Infection of primary human tonsil T cells with the cluster gene mutants.
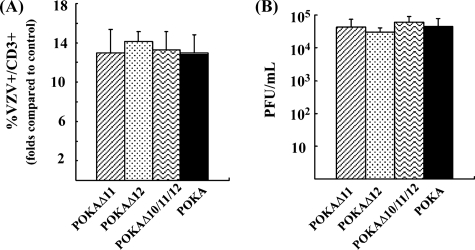
Tonsil T cells were collected 48 h after incubation on HELF cell monolayers infected with POKA or the cluster gene mutants, including POKAΔ11, POKAΔ12, and POKAΔ10/11/12. The T cells were stained with anti-CD3 antibody as a T-cell marker and with polyclonal anti-VZV human IgG to detect VZV protein expression and analyzed by flow cytometry. Our previous experiments demonstrated that deleting ORF10 did not impair VZV T cell tropism (3). These experiments showed that deleting POKAΔ11, POKAΔ12, or POKAΔ10/11/12 also had no significant effect on levels of T-cell infection. The percentages of CD3 and VZV double-positive T cells were similar for all viruses tested compared to those for the mock-infected T-cell control (Fig. 4A). In addition, the titers of infectious virus in the HELF cell monolayers were determined after harvesting the T cells. The titers were similar in all preparations (Fig. 4B), indicating that the cluster gene mutants replicated as well as POKA in HELF cells and that the conditions for the T-cell infection assay were consistent for POKA and the cluster gene mutants.
FIG. 4.
Replication of POKA and the ORF10-to-ORF12 cluster gene mutants in human tonsil T cells. Primary human tonsil T cells were infected by incubation with HELF cell-infected monolayers. At 48 h postinfection, T cells were collected and stained with anti-CD3 antibody and polyclonal anti-VZV human IgG and analyzed by flow cytometry. (A) T-cell flow cytometry assay. The bars represent the folds of CD3 and VZV double-positive T cells detected for T cells incubated with POKA or the mutant viruses in comparison to values for the uninfected T-cell control. The error bar indicates the standard deviation. (B) Titrations of HELF cell-infected monolayers. Virus titers were determined by an infectious focus assay using melanoma cells. Each bar represents the mean titer from three independent wells, and the error bars represent the standard deviations.
Effects of cluster gene deletions on VZV virion morphogenesis in vitro.
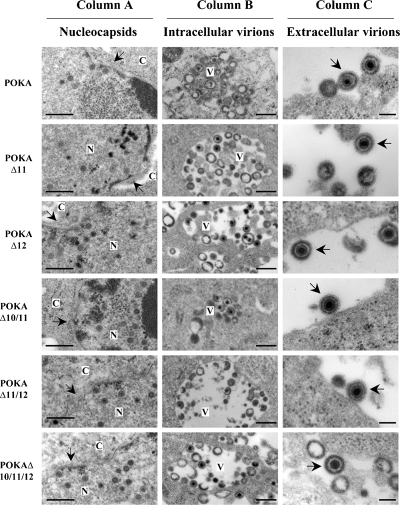
Growth kinetics and plaque morphologies of VZV mutants can be indistinguishable even when virion assembly is severely disrupted, as demonstrated by our analysis of ORF47 kinase-deficient mutants (1). To investigate the effect of deleting the cluster genes, including ORF11, ORF12, ORF10 and -11, ORF11 and -12, or ORF10 to -12, singly or in combination, on the virion assembly process, melanoma cells infected with POKA or the cluster gene mutants were fixed 48 h postinfection and examined by TEM. Single or multiple deletions of the cluster genes had no detectable effect on VZV virion assembly (Fig. 5). As we reported previously, the ORF10 single deletion also had no apparent effect on virion morphogenesis and trafficking in infected cells (3). Melanoma cells infected with POKA or the cluster gene mutants showed abundant nucleocapsids, the majority of which appeared to contain viral DNA (Fig. 5, column A). Many intracellular virions were observed in cytoplasmic vacuoles in cells infected with POKA and cluster gene mutants (Fig. 5, column B). Interestingly, there was no apparent difference in the densities of the tegument layer in viral particles observed in the cytoplasm of cells infected with single, double, or triple deletion mutants in comparison to wild-type POKA, even though ORF10, -11, and -12 are predicted to be major tegument proteins. Virion structures were the same as those observed for POKA-infected cells, including the DNA core, electron-dense tegument material, and the typical glycoprotein projections on their envelopes (Fig. 5, column C).
FIG. 5.
Effects of cluster gene deletions on VZV virion morphogenesis in vitro. Melanoma cells were fixed and examined by TEM at 48 h postinfection with POKA and the ORF10-to-ORF12 cluster gene mutants. The viruses tested are indicated at the left. Column A shows nucleocapsid formation in nuclei; the arrows point to the nuclear membranes. Column B shows virions in cytoplasmic vacuoles. Column C shows the structure of extracellular virions; mature virus particles with glycoprotein envelope projections are indicated by arrows. Bars, 0.5 μm (A and B) and 0.2 μm (C). N, nucleus; C, cytoplasm; V, vacuoles.
Replication of the ORF10-to-ORF12 cluster gene mutants in skin xenografts in vivo.
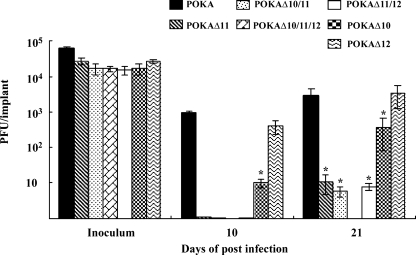
In these experiments, skin xenografts in SCID mice were infected with POKA or the cluster gene mutants. Infected xenografts were harvested at 10 and 21 days after infection, and their virus yields were determined on melanoma cell monolayers by an infectious center assay. The growth characteristics of POKAΔ12 in skin xenografts were similar to those of POKA, with mean titers of ∼1 × 103 and ∼6 × 103 PFU per xenograft at 10 and 21 days after infection (Fig. 6). As we have shown previously, POKAΔ10 exhibited up to a 10-fold decrease in replication in skin compared to that for POKA (3) (Fig. 6). The effect of deleting ORF11 was even more striking. POKAΔ11 was recovered from only one of six xenografts at 10 days postinfection, with only a few plaques visible in the monolayer; virus was isolated from two of six skin xenografts, with a mean titer of only ∼1 × 101 PFU per xenograft at 21 days postinfection. The combined deletion of ORF10 and ORF11 yielded virus from two of six implants, which was similar to the replication efficiency of POKAΔ11. In addition, whereas the ORF12 deletion had no effect alone, the removal of both ORF11 and ORF12 severely affected virus replication in skin. Only two of five infected xenografts yielded infectious virus; the virus growth was similar to that for POKAΔ11 and POKAΔ10/11, showing about a 500-fold reduction in virus yield at 21 days postinfection compared to POKA (Fig. 6). Furthermore, no virus was recovered from any of the xenografts infected with the triple-deletion mutant, POKAΔ10/11/12, at either 10 or 21 days after infection. To ensure that this block in skin infectivity was associated with deletion of the ORF10/11/12 gene cluster, these experiments were repeated, and the results were found to be reproducible.
FIG. 6.
Replication of the ORF10-to-ORF12 cluster gene mutants in human skin xenografts in SCID-hu mice in vivo. Skin xenografts were infected with POKA, POKAΔ10, POKAΔ11, POKAΔ12, POKAΔ10/11, POKAΔ11/12, or POKAΔ10/11/12. Xenografts were harvested at 10 and 21 days after inoculation, and infectious virus yields of each sample were determined by infectious focus assays. Each bar represents the mean viral titer, and error bars represent the standard deviations. The asterisks indicate significant differences in viral titers (P < 0.05) compared to those for POKA.
Since infection with POKAΔ10 was associated with the accumulation of unenveloped capsids with aggregates of electron-dense tegument-like proteins in the cytoplasm of infected skin cells (3), we analyzed skin xenografts infected with the cluster gene mutants for changes in virion assembly. POKAΔ12 virus particles were easily located in skin cells and did not differ from POKA virions (data not shown). Virus particles were not identifiable in skin xenografts infected with POKAΔ11, POKAΔ10/11, and POKAΔ11/12 because of the severe reduction in replication.
Effects of the cluster gene deletions on VZV lesion formation in skin xenografts in vivo.
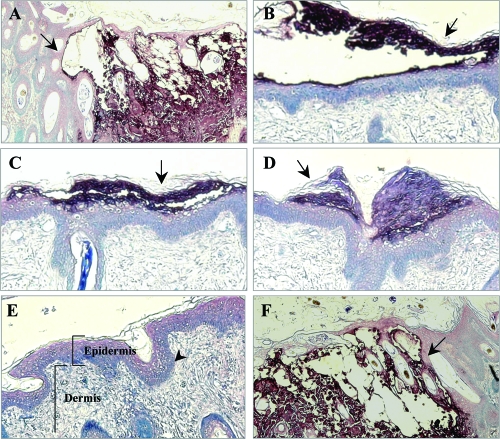
To examine effects of cluster gene deletions on lesion formation, sections from skin xenografts harvested at 21 days after infection were stained with human polyclonal anti-VZV IgG and biotinylated secondary antibody. Lesion formation by POKAΔ12 was similar to POKA (Fig. 7, panels A and F). These viruses produced large necrotic lesions in the epidermis and penetrated across the basement membrane, extending into the dermal layer. In contrast, only small lesions expressing VZV proteins that were restricted to the epidermal layer were observed for skin xenografts infected with POKAΔ11, POKAΔ10/11, or POKAΔ11/12 (Fig. 7, panels B and D). In skin xenografts infected with POKAΔ10/11/12, the structure of the epidermis and dermis remained intact and no lesions were detected (Fig. 7, panel E). These observations of reduced cell-cell spread in skin were consistent with the limited replication of these viruses shown by infectious focus assay.
FIG. 7.
Lesion formation in skin xenografts infected with ORF10-to-ORF12 cluster gene mutants. Deparaffinized skin sections harvested at days 21 after infection were incubated with human polyclonal anti-VZV IgG and stained with biotinylated secondary antibody (purple; magnification, ×10). Sections representative of skin xenografts infected with each virus are shown. Large lesions were observed for skin infected with POKA (A) and POKAΔ12 (F). Small lesions (arrows) restricted to the epidermal layer were observed for skin infected with POKAΔ11 (B), POKAΔ10/11 (C), and POKAΔ11/12 (D). POKAΔ10/11/12 (E) did not produce detectable skin lesions. The normal structures of the epidermis, the dermis, and the basement membrane that separates the epidermis from the underlying dermis are indicated in this section.
DISCUSSION
The VZV ORF9-to-ORF12 gene cluster is conserved in most alphaherpesviruses, in which it is designated UL46 to UL49. Deletion of ORF9 was incompatible with VZV replication in these experiments using mutagenesis of POKA cosmids, which is consistent with the report by Tischer et al. showing that ORF9 was essential by using a POKA BAC system (39). Inactivating the ORF9 start codon in an infectious POKA BAC clone blocked virus propagation except with baculovirus ORF9 complementation, and repairing the mutation resulted in growth kinetics equivalent to those of the POKA BAC. In our cosmid experiments, VZV replication was restored when ORF9 was expressed from a nonnative site in the short unique region of the VZV genome. Thus, like its homologue in Marek's disease virus (11), VZV ORF9 appears to be an essential gene. In contrast, deleting PRV UL49 had no apparent effect on virus growth in cultured cells or in a rat model (10). The consequences of deleting UL49 for HSV-1 and bovine herpesvirus (BHV) type 1 replication were significant, but UL49 remained dispensable (13, 14, 26, 34). The differential requirement for ORF9 and HSV-1 UL49 (VP22) is not unexpected, since these proteins share only 25% identity (4). Taken together, these and previous analyses of the ORF9-to-ORF12 gene cluster and the HSV-1 UL46-to-UL49 homologues show that whereas ORF9 is essential for VZV replication and ORF10 is dispensable, HSV-1 UL48 (VP16) is essential and UL49 (VP22) is not required.
Further studies of ORF9 by immuno-EM showed that ORF9, like its HSV and PRV homologues (20, 31), is absent from perinuclear virions and is incorporated into mature virions during secondary envelopment in the cytoplasm. These experiments showed that ORF9 is a tegument protein and that it is abundant in extracellular virions. ORF9 was also found in association with aggregates of electron-dense materials in the nuclei of VZV-infected cells, whereas Cilloniz et al. (4) detected ORF9 exclusively in the cytoplasm by confocal microscopy. The nuclear localization of ORF9 homologues appears to be conserved among the different alphaherpesvirus, including HSV, PRV, MDV, and BHV, although the biological function that ORF9 may have at these nuclear sites is not known (10, 12, 26, 33). In addition, we found that VZV ORF9 in infected cell extracts was resolved into three differently migrating forms in denaturing gel electrophoresis. Of interest, HSV-1 VP22 is also present as three forms in infected cell lysates; only the higher-molecular-weight form was packaged into infectious virions (2). At this time, it is unknown whether one form of VZV ORF9 is packaged into virions while the other forms are not; it is possible that the ORF9 multiple forms result from different levels of phosphorylation, which may contribute to the incorporation of virions.
Based on coimmunoprecipitation experiments, these experiments demonstrated that VZV ORF9 binds to the major VZV glycoprotein, gE, suggesting that ORF9 and gE may form a complex in VZV-infected cells. Cilloniz et al. recently demonstrated that ORF9 protein interacts with IE62 and also with β-tubulin (4). Their observations suggest that ORF9 may attach to microtubules, playing a role in the intracellular transport of nonenveloped virions, and that ORF9 may facilitate IE62 incorporation into the virion tegument, either before or after ORF9 is bound to microtubules. However, β-tubulin did not bind to gE, based on immunoprecipitation with the antibody to β-tubulin antibody, indicating that gE is not part of a complex between ORF9, IE62, and β-tubulin. Thus, the formation of ORF9 and gE complexes that we observed is likely to occur under other circumstances in the infected cell. Our immuno-EM analysis of ORF9 showed that ORF9 is present in the putative trans-Golgi-derived membranes, the area where VZV gE is known to be located during secondary envelopment (19). It has been suggested that the formation of mature virions requires protein-protein interactions among tegument components and between tegument proteins and glycoproteins present in the secondary envelope (28). UL49, the ORF9 homologue in HSV and PRV, has been shown to form complexes with several glycoproteins, including gE, gD, and gM, which have been implicated in secondary envelopment (15, 17). ORF9 is likely to be essential because it has critical functions at different steps of VZV virion assembly through its interactions with gE as well as IE62, both of which are also required for VZV replication.
In contrast to ORF9, VZV mutants with deletions of ORF11 and ORF12 replicated as well as POKA in melanoma cells and human fibroblasts in vitro, and as we observed when ORF10 was deleted from POKA (3), removal of these genes did not interfere with the transfer of POKAΔ11 and POKAΔ12 viruses from fibroblasts into primary human tonsil T cells in vitro. Furthermore, the three nonessential genes ORF10, ORF11, and ORF12 could be deleted in pairs or as a triple deletion without any consequences for VZV growth kinetics or VZV virion formation and maturation, compared to POKA in cultured cells. The effects of deleting some combinations of genes related to ORF10, ORF11, and ORF12 from other alphaherpesviruses have also been described. Removing UL46 and UL47 from HSV-1, MDV, and PRV impaired viral replication moderately (11, 24, 42, 43), whereas deleting UL48 from PRV and MDV caused significant reductions (11, 18). PRV could be propagated when all four genes, UL46 to UL49, were absent, but this mutant exhibited severely impaired replication, and virions lacked the electron-dense tegument layer and had no visible glycoprotein spikes when secondary envelopment was detected (16). Given these observations, the fact that POKAΔ10/11/12 showed no evidence of defective virion assembly or any reduced density of tegument structures by EM was unexpected, suggesting that the missing teguments might be replaced by either viral or cellular proteins within the tegument. VZV appears to be unusual in that the functions of ORF10, ORF11, and ORF12 are redundant and that the absence of all three can be fully compensated for during VZV replication in vitro.
As reported previously, ORF10 is necessary for optimal VZV replication in skin xenografts in vivo (3). This investigation of ORF11 and ORF12 gene functions during the pathogenesis of skin infection using the POKAΔ11 and POKAΔ12 mutants demonstrated that ORF12 was dispensable, whereas ORF11 was an important VZV virulence determinant for skin. At this point, the functions of ORF12 remain undefined both in vitro and in vivo. Information about the functions of the HSV homologue UL46 (VP11/12) is also limited. There is some evidence to suggest that UL46 may be involved in the modulation of UL48 (VP16) activity in transactivating α genes (27, 42, 43). In contrast to the deletion of ORF12, the deletion of ORF11 had an impact on VZV virulence in skin that was even more significant than that seen when ORF10 was removed. Without ORF11, the frequency with which any infectious virus was recovered from skin xenografts was reduced dramatically, and peak VZV titers were approximately 700-fold and 500-fold lower than those for POKA and POKAΔ12 at 10 and 21 days after inoculation. If skin lesions were formed, they were very small, and cytopathic changes were restricted to the epidermis rather than extending across the basement membrane into the dermis, which is a marker of substantially impaired virulence. The consequences of removing ORF10 and ORF11 or ORF11 and ORF12 in combination were not more significant than those associated with removing ORF11 alone. However, a role for ORF12 may be suggested by the fact that removing this gene along with ORF10 and ORF11, both of which affect the pathogenesis of VZV infection in skin, resulted in a complete inhibition of infectivity. This observation suggests that compensating mechanisms, which permit some replication when ORF10 and ORF11 are deleted, do not overcome the incremental effect of deleting ORF12 from the VZV genome.
Although no consequences were observed in vitro, the effects of deleting ORF10 were demonstrated by an EM analysis of VZV-infected skin xenografts, which showed impaired secondary envelopment of POKAΔ10 virions compared to that seen for POKA (3). The absence of ORF10 was also associated with the formation of many aggregates of capsids together with tegument-like materials in the cytoplasm of epidermal cells in vivo, which may be linked to defective secondary envelopment. Comparable studies of skin xenografts infected with POKAΔ11 were not possible, because its replication was even more defective than that of POKAΔ10, and no virions were detected in skin infected with this deletion mutant. In PRV, the UL47 deletion impairs virus maturation and egress (24). It is shown that HSV UL47 is one of the most abundant tegument proteins and is present at the outer layer of virion tegument; this protein has the capacity to enhance UL48 (VP16) expression in vitro (27, 42, 43). UL47 has also been characterized as a nucleocytoplasmic shuttling protein for both HSV and BHV (40, 44). These observations suggest that VZV ORF11 may share functions with their homologue, including an influence on the expression of the major VZV transactivating proteins, a role in virion assembly, or functions related to the binding and export of RNAs from the nucleus. While its functions are completely redundant in cultured cells, it is possible that ORF11 influences the transactivation of VZV genes and has functions in virion assembly or RNA transportation that are important and irreplaceable for replication in skin in vivo.
In summary, with the exception of ORF9, the individual genes within the ORF9-to-ORF12 gene cluster are dispensable and can be deleted simultaneously without any apparent effect on VZV replication in vitro. However, ORF11 was revealed to be an important virulence determinant when the POKAΔ11 mutant was challenged to replicate in differentiated human skin cells in vivo, and the ORF10-to-ORF12 cluster was essential for the pathogenesis of VZV skin infection.
Acknowledgments
This work was supported by NIH grants AI053846 and AI20459.
We thank Nafisa Ghori and Linda Lew Yasukawa for their valuable technical assistance.
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.Besser, J., M. Ikoma, K. Fabel, M. H. Sommer, L. Zerboni, C. Grose, and A. M. Arvin. 2004. Differential requirement for cell fusion and virion formation in the pathogenesis of varicella-zoster virus infection in skin and T cells. J. Virol. 7813293-13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaho, J. A., C. Mitchell, and B. Roizman. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J. Biol. Chem. 26917401-17410. [PubMed] [Google Scholar]
- 3.Che, X., L. Zerboni, M. H. Sommer, and A. M. Arvin. 2006. Varicella-zoster virus open reading frame 10 is a virulence determinant in skin cells but not in T cells in vivo. J. Virol. 803238-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cilloniz, C., W. Jackson, C. Grose, D. Czechowski, J. Hay, and W. T. Ruyechan. 2007. The varicella-zoster virus (VZV) ORF9 protein interacts with the IE62 major VZV transactivator. J. Virol. 81761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, J. I., and K. E. Seidel. 1994. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J. Virol. 687850-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, J. I., S. E. Straus, and A. M. Arvin. 2007. Varicella-zoster virus replication, pathogenesis, and management, p. 2773-2818. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Cohrs, R. J., M. Barbour, and D. H. Gilden. 1996. Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J. Virol. 702789-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davision, A. J. 2000. Molecular evolution of alphaherpesviruses. p 25-50. In A. M. Arvin and A. A. Gershon (ed.), Varicella zoster virus virology and clinical management. Cambridge University Press, Cambridge, United Kingdom.
- 9.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 671759-1816. [DOI] [PubMed] [Google Scholar]
- 10.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 761959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorange, F., S. El Mehdaoui, C. Pichon, P. Coursaget, and J. F. Vautherot. 2000. Marek's disease virus (MDV) homologues of herpes simplex virus type 1 UL49 (VP22) and UL48 (VP16) genes: high-level expression and characterization of MDV-1 VP22 and VP16. J. Gen. Virol. 812219-2230. [DOI] [PubMed] [Google Scholar]
- 13.Duffy, C., J. H. Lavail, A. N. Tauscher, E. G. Wills, J. A. Blaho, and J. D. Baines. 2006. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J. Virol. 808664-8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, G., W. Hafezi, A. Whiteley, and E. Bernard. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 799735-9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farnsworth, W., T. W. Wisner, and D. C. Johnson. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, W., H. Granzow, and T. C. Mettenleiter. 2003. A pseudorabies virus recombinant simultaneously lacking the major tegument proteins encoded by the UL46, UL47, UL48, and UL49 genes is viable in cultured cells. J. Virol. 7712891-12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 768208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, W., B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 766729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gershon, A. A., Z. Zhu, D. L. Sherman, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 686372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J. Virol. 781314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grose, C., D. M. Perrotta, and P. A. Brunell. 1979. Cell-free varicella-zoster virus in cultured human melanoma cells. J. Gen. Virol. 4315-27. [DOI] [PubMed] [Google Scholar]
- 22.Kinchington, P. R., D. Bookey, and S. E. Turse. 1995. The transcriptional regulatory proteins encoded by varicella-zoster virus open reading frames (ORFs) 4 and 63, but not ORF 61, are associated with purified virus particles. J. Virol. 694274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinchington, P. R., J. K. Hougland, A. M. Arvin, W. T. Ruyechan, and J. Hay. 1992. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J. Virol. 66359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 768820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ku, C. C., J. A. Padilla, C. Grose, E. C. Butcher, and A. M. Arvin. 2002. Tropism of varicella-zoster virus for human tonsillar CD4+ T lymphocytes that express activation, memory, and skin homing markers. J. Virol. 7611425-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, X., B. Chow, Y. Li, C. Raggo, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J. Virol. 693863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKnight, J., P. Pellett, F. Jenkins, and B. Roizman. 1987. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate α-trans-inducing factor-dependent activation of α genes. J. Virol. 61992-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffat, J. F., M. D. Stein, H. Kaneshima, and A. M. Arvin. 1995. Tropism of varicella-zoster virus for human CD4+ and CD8+ T lymphocytes and epidermal cells in SCID-hu mice. J. Virol. 695236-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moriuchi, H., M. Moriuchi, S. E. Straus, and J. I. Cohen. 1993. Varicella-zoster virus open reading frame 10 protein, the herpes simplex virus VP16 homolog, transactivates herpesvirus immediate-early gene promoters. J. Virol. 672739-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naldinho-Souto, R., B. Helena, and T. Minson. 2006. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J. Virol. 802582-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niizuma, T., L. Zerboni, M. H. Sommer, H. Ito, S. Hinchliffe, and A. M. Arvin. 2003. Construction of varicella-zoster virus recombinants from parent Oka cosmids and demonstration that ORF65 protein is dispensable for infection of human skin and T cells in the SCID-hu mouse model. J. Virol. 776062-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 736769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 7410041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross, J., M. Williams, and J. I. Cohen. 1997. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology 234186-195. [DOI] [PubMed] [Google Scholar]
- 36.Santos, R. A., C. C. Hatfield, N. L. Cole, J. A. Padilla, J. F. Moffat, A. M. Arvin, W. T. Ruyechan, J. Hay, and C. Grose. 2000. Varicella-zoster virus gE escape mutant VZV-MSP exhibits an accelerated cell-to-cell spread phenotype in both infected cell cultures and SCID-hu mice. Virology 275306-317. [DOI] [PubMed] [Google Scholar]
- 37.Spengler, M., N. Niesen, C. Grose, W. T. Ruyechan, and J. Hay. 2001. Interactions among structural proteins of varicella zoster virus. Arch. Virol. Suppl. 1771-79. [DOI] [PubMed] [Google Scholar]
- 38.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, NY.
- 39.Tischer, B. K., B. B. Kaufer, M. Sommer, F. Wussow, A. M. Arvin, and N. Osterrieder. 2007. A self-excisable infectious bacterial artificial chromosome clone of varicella-zoster virus allows analysis of the essential tegument protein encoded by ORF9. J. Virol. 8113200-13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verhagen, J., M. Donnelly, and G. Elliott. 2006. Characterization of a novel transferable CRM-1-independent nuclear export signal in a herpesvirus tegument protein that shuttles between the nucleus and cytoplasm. J. Virol. 8010021-10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Y., and J. McKnight. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 671482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, Y., D. A. Sirko, and J. L. C. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng, C., R. Brownlie, L. A. Babiuk, and S. van Drunen Littel-van den Hurk. 2004. Characterization of nuclear localization and export signals of the major tegument protein VP8 of bovine herpesvirus-1. Virology 324327-339. [DOI] [PubMed] [Google Scholar]