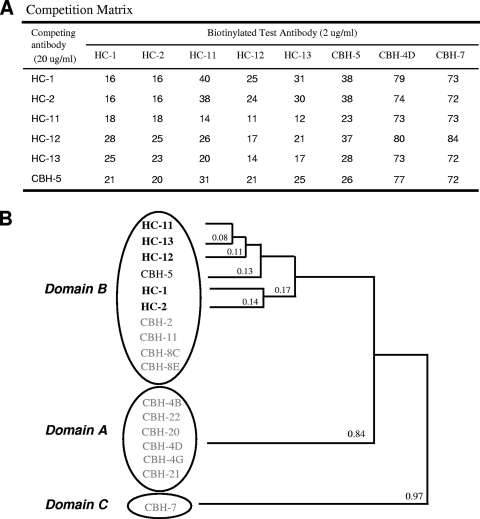
FIG. 2.
Analysis of competition between the five HC HMAbs (bold type) and other HCV HMAbs to domain B. (A) The cross-competition matrix is based on the binding of a biotinylated test antibody to genotype 1b HCV E2 (expressed in 293T cells), captured onto Galanthus nivalis lectin-coated microtiter plates in the presence of various concentrations of competing antibodies (12). The bound biotinylated test antibody was detected by using alkaline phosphatase-conjugated streptavidin and p-nitrophenyl phosphate as the substrate. The numbers shown represent the percentages of residual binding. (B) Hierarchical clustering of HCV HMAbs based on relatedness in a competitive binding assay. Solid lines with numbers indicate the relatedness of the two adjoining antibodies. In this analysis, the cross-competition percentages of any two antibodies were averaged and expressed as a fraction; the smaller the fraction, the greater the cross-competition (12). Circles are clusters of antibodies in a specific domain. Three domains are indicated at the left. The relatedness of domain A and C HMAbs has been described previously (12).