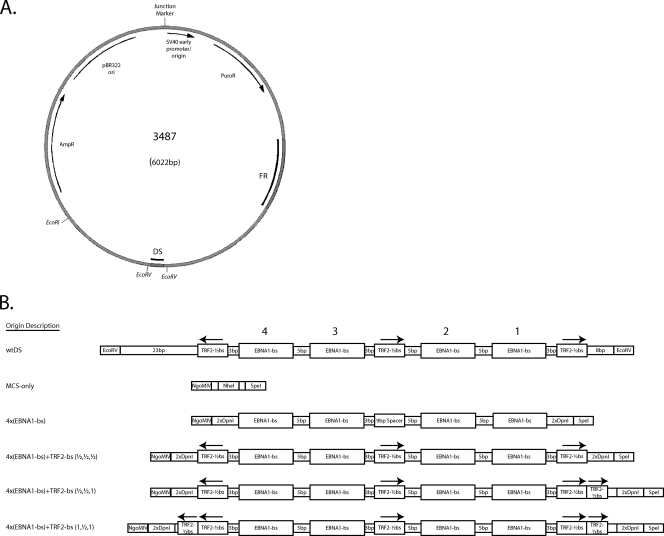
FIG. 1.
oriP plasmid backbone used to introduce the MCS and engineered origins of DNA synthesis tested in the present study. (A) The plasmid p3487 is derived from the pPUR vector (Clontech), which contains a pBR322 origin for plasmid replication in Escherichia coli and resistance to both ampicillin and puromycin. The wild-type oriP sequence has been inserted into a unique BamHI restriction site, which was destroyed near FR but was retained near DS. The MCS of plasmid p3488 was inserted between the EcoRV sites flanking DS. Subsequent insertions of the engineered origins, or the reintroduction of wtDS, were made into unique NgoMIV and SpeI sites present within this MCS. (B) Graphical representation of each class of origin tested. Within each class, only the sequences corresponding to the EBNA1-binding sites differ. Plasmids p3487 and p3567, which includes 1.2 kb of lambda phage DNA inserted in the EcoRI site of p3487, contain the wtDS element between the EcoRV sites as found in the B95-8 strain of EBV (4) and serve as positive-control origins. Plasmid p3488 has a 22-bp MCS inserted between the EcoRV sites of p3487 to remove and replace wtDS (MCS-only) and serves as an origin-less negative control. Test plasmids contain four EBNA1-binding sites of a range of different affinities in the same arrangement observed in wtDS, in the absence or presence of TRF2-half-binding sites. Two origins (wtDS and 4×FRbs) were further modified to include one or two complete TRF2-binding sites as well. The arrows above the TRF2-binding sites denote the N-to-C-terminal orientation that TRF2 would use to bind to these sites, as are found in wtDS. All origins were inserted between the NgoMIV and SpeI sites of p3488 to ensure that the only differences between the test plasmids were the origin sequences. The sequences of all origins tested are given in Fig. S1 in the supplemental material.