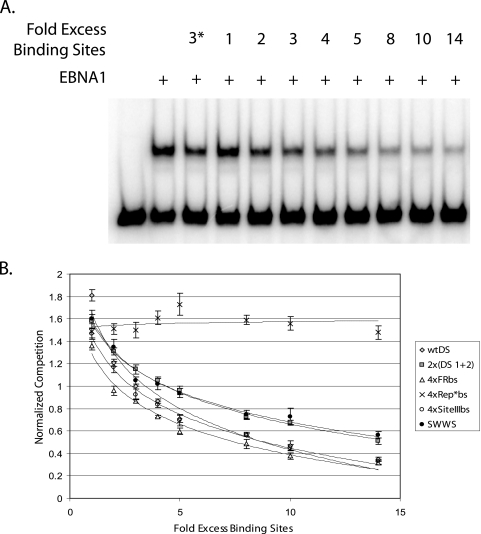
FIG. 2.
Rank order of the affinity of EBNA1 for its binding sites in origins of DNA synthesis as determined by competitive EMSA. (A) A total of 5 fmol of end-labeled probe containing one EBNA1-binding site from FR was incubated either in the absence or in the presence of 1.6 nM dnEBNA1/Softag1 in a reaction volume of 25 μl at room temperature for 30 min. Reactions contained increasing amounts (1- to 14-fold excess of binding sites) of unlabeled DNA fragments containing either wtDS or one of the engineered origins of DNA synthesis. A threefold excess of wtDS EBNA1-binding sites was included in one lane on all gels, marked with an asterisk, as an unlabeled competitor control and was used for sample normalization across experiments. Samples were electrophoresed through a 4% polyacrylamide gel at 300 V at 4°C for 1.5 to 2 h, dried onto Whatman paper, exposed to a storage phosphor screen, and visualized by scanning on a Storm 640 PhosphorImager. (B) The amount of competition for binding by EBNA1 produced from each origin tested was normalized to the amount of competition for binding by EBNA1 produced from a threefold excess of wtDS EBNA1-binding sites. These values were plotted on the y axis against the fold excess of unlabeled EBNA1-binding sites added to each reaction in order to determine the rank order of the affinity of EBNA1 for all origins tested. A 25-fold excess in the binding sites of the 4×Rep*bs containing DNA fragment was required to produce an amount of competition similar to that of 3-fold excess wtDS EBNA1-binding sites (data not shown). The rank order is 4×FRbs > wtDS ≈ 4×SiteIIIbs > 2×(DS1+2) ≈ S-W-W-S ≫ 4×Rep*bs, as determined by experiments conducted in triplicate.