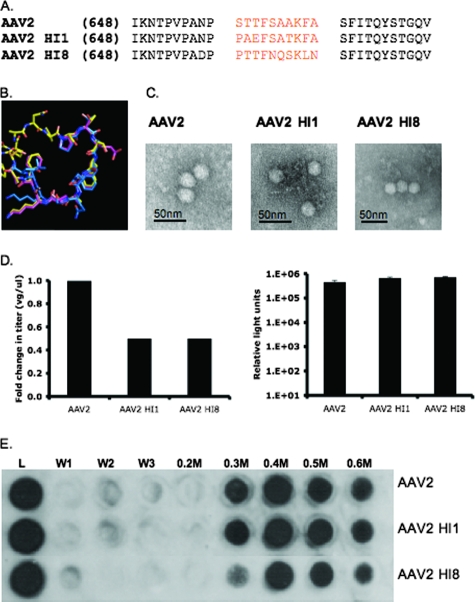
FIG. 3.
AAV2 HI1 and AAV2 HI8 substitution mutant characterization. (A) Sequence alignment of AAV2, AAV1, and AAV8 HI loop amino acids. (B) Structural alignment of AAV2, AAV1, and AAV8 HI loops (yellow, pink, and blue). (C) Electron micrograph images of AAV2, AAV2 HI1, and AAV2 HI8 1× PBS-Ca2+-Mg2+-dialyzed, CsCl-purified (previously mentioned), DNA-containing particles. (D) Radioactive DNA dot blotting shown as relative change in titer (left side) and luciferase assay on infected 293 cells with 3,000 VGs/cell (right side; n = 3; standard deviation = black bars). (E) Heparin binding profiles of AAV2, AAV2 HI1, and AAV2 HI8. A 500-μl volume of heparin type IIIS-conjugated agarose beads (Sigma) was incubated with 1010 VG-containing particles (L = load) for 10 min at RT. The washes (1× PBS) and elutions (0.2 to 0.6 M PBS) were collected, and capsids were detected via Western dot blot analysis with monoclonal antibody A20 (1:20).