Abstract
Limited cell ingrowth is a major problem for tissue engineering and the clinical application of porous biomaterials as bone substitutes. As a first step, migration and proliferation of an interacting cell population can be studied in two-dimensional culture. Mathematical modelling is essential to generalize the results of these experiments and to derive the intrinsic parameters that can be used for predictions. However, a more thorough evaluation of theoretical models is hampered by limited experimental observations. In this study, experiments and image analysis methods were developed to provide a detailed spatial and temporal picture of how cell distributions evolve. These methods were used to quantify the migration and proliferation of skeletal cell types including MG63 and human bone marrow stromal cells (HBMSCs). The high level of detail with which the cell distributions were mapped enabled a precise assessment of the correspondence between experimental results and theoretical model predictions. This analysis revealed that the standard Fisher equation is appropriate for describing the migration behaviour of the HBMSC population, while for the MG63 cells a sharp front model is more appropriate. In combination with experiments, this type of mathematical model will prove useful in predicting cell ingrowth and improving strategies and control of skeletal tissue regeneration.
Keywords: skeletal tissue engineering, cell migration, image analysis, mathematical modelling
1. Introduction
Insufficient tissue formation is a major problem in the clinical application of porous biomaterials as bone substitutes in vivo and the generation of functional tissue engineered constructs in vitro (El-Ghannam 2005; Mauney et al. 2005; Mistry & Mikos 2005). Typically, peripheral tissue formation can be observed, while central zones within a construct remain void (Bancroft et al. 2002; Erli et al. 2006). Such spatially inhomogeneous tissue formation can be attributed to inadequate nutrient transport, but may also result from limited cell ingrowth following non-uniform seeding. Cell ingrowth may be hampered by pore size restrictions, unfavourable surface characteristics, or may be counteracted by chemotactic movement towards external supplies of nutrients and other important biochemical factors. To promote ingrowth, cell attachment and mobility can be optimized by applying surface modifications to the biomaterial (Dimilla et al. 1991; Lauffenburger & Horwitz 1996; Dee et al. 1999; Erli et al. 2006). As a first step in a systematic approach, cell migration can be studied in two-dimensional culture, combining ease of observation and a high level of control over the cellular environment, although, clearly, important differences between two- and three-dimensional cell adhesion exist (Cukierman et al. 2002).
In general, two experimental approaches can be distinguished, whose results can be related theoretically (Farrell et al. 1990). In the first approach, time-lapse video and image analysis software are used to track the movement of individual cells that are sparsely distributed (Krooshoop et al. 2003). In the second approach, migration of an interacting population of cells is monitored. For example, an assay used in the study of wound healing is based on the removal of a band of cells from a monolayer by scraping, after which the rate at which the cells repopulate the area is recorded (Maini et al. 2004a). Similarly, cells can be seeded in a circle that expands as the cell population spreads further out (Dixit et al. 2001; Shin et al. 2004; Dolle et al. 2005). Mathematical modelling is essential to interpret and generalize the results of these experiments and to derive the intrinsic parameters that can be used to predict cell behaviour (Tranquillo et al. 1988; Murray 2002). For example, using theoretical modelling analogous to diffusion, the random motility coefficient of an osteoblast population was derived from the position of the cell front in an under-agarose assay, on surfaces modified with adhesive peptides (Dee et al. 1999).
Fisher's equation has been used to represent experimental data from wound-healing assays (Takamizawa et al. 1996; Savla et al. 2004). In the context of tissue engineering, Fisher's equation has been applied to a wound-healing migration assay for human peritoneal mesothelial cells (Maini et al. 2004a,b). This equation describes the behaviour of a cell population as a combination of random cell motion and logistic proliferation, i.e. proliferation up to a maximum cell density. Fisher's equation predicts that, under certain conditions, the cell migration front takes the form of a travelling wave of fixed shape that propagates at a constant speed (Sherratt & Marchant 1996; Murray 2002; Maini et al. 2004b). This velocity is dependent only on a simple combination of the migration coefficient and the proliferation rate. Therefore, once the migration front velocity has been determined experimentally, it is straightforward to derive the migration coefficient if the proliferation rate is known.
Measuring the position of the cell migration front is relatively straightforward in cases where a sharp front exists. However, for a more disperse front the definition is less clear. The front definition can, for example, be based on a certain cell density level to be reached, which is unambiguous in cases where the front has a constant shape that travels at a certain speed, but introduces ambiguity in the early phase while the front shape is still developing from the initial conditions. It is therefore important not to rely on limited observations, but to obtain a complete picture of the spatial distribution of cells as a function of time.
Furthermore, it has been observed that the migration front can be relatively sharp while Fisher's equation yields a comparatively smooth solution (Maini et al. 2004a; Callaghan et al. 2006). This raises the issue as to what extent this modelling approach is appropriate. Thus, a ‘sharp front’ version of Fisher's equation has been proposed, but deemed unnecessary, as it was possible to obtain a good fit of the front velocity with the standard Fisher model (Maini et al. 2004a). However, instead of a largely phenomenological description of the front velocity, a correct representation of the actual cell distribution is needed to indicate that the proposed model captures the main underlying mechanisms involved. From a tissue engineering perspective, it is important to consider whether mitosis drives cell migration or vice versa (Murray 2003, p. 450). In other words, do the cells migrate outwards, which gives them room to divide, or are they pushed forward as a result of cell division and which mechanism is more important (Simpson et al. 2007)?
The objective of the present study was a detailed, quantified characterization of both the migration and proliferation behaviour of osteoblast-like MG63 and human bone marrow stromal cell (HBMSC) populations, which are relevant in the study and application of skeletal regeneration. This study set out to develop experimental methods for measuring the evolving cell density distribution of these cell types at high spatial and temporal resolution. These data then provided a more thorough evaluation of the ability of mathematical models, like Fisher's equation, to represent the cell population behaviour and, critically, insight into whether the underlying assumptions on cell behaviour are appropriate and can be applied to cell adhesion, spreading and migration in skeletal tissue regeneration strategies.
2. Methods
2.1 Experimental materials and methods
Experiments were performed for both the human osteosarcoma MG63 cell line and passage zero HBMSCs. All tissue culture reagents were purchased from Sigma-Aldrich, UK unless stated otherwise. MG63 culture medium consisted of Dulbecco's modified Eagle's medium (DMEM, 91%) and foetal calf serum (FCS, 9%). HBMSC culture medium consisted of alpha-modified Eagle's medium (αMEM, 89%), Penicillin–Streptomycin (50×, 2%; Cambrex Bio Science, UK) and FCS (9%). The bone marrow sample was obtained from a haematologically normal patient (female 79 years old, F79) undergoing routine total hip replacement surgery, with approval from the Southampton Hospital Ethics Committee and appropriate patient consent. Briefly, HBMSCs were harvested, centrifuged at 1100 r.p.m. for 4 min, resuspended in αMEM and passed through a 70 μm cell strainer. HBMSCs were cultured for 6 days in culture medium in a humidified incubator at 37°C and 5% CO2, after which the cells were washed twice with phosphate-buffered saline (PBS) and expanded for another 6 days.
2.2 Circle migration assay
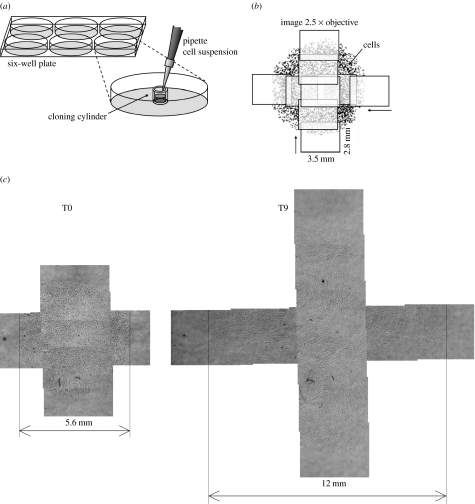
The cells were seeded in a circle using the following procedure (figure 1a): cloning cylinders (n=6, polystyrene, Sigma-Aldrich, UK, Cat. No. C7983) were positioned in one or two six-well plates. Cells were detached using trypsin and 50 μl of 1.7×105 cells ml−1 cell suspension was added to each cylinder, which corresponds to 50 000 cells cm−2 based on a 4.7 mm internal diameter. After allowing 4 h for attachment, medium was added and the cylinder removed, resulting in a circle of cells. To monitor the spreading of this cell circle, the wells were photographed immediately afterwards and every 24 h over the next 9 days at low magnification (2.5×), with a Zeiss Axiovert 200 microscope fitted with an AxioCam CCD camera, using Axiovision v. 4.5 software. During photography, the plate was positioned at a heated stage kept at 37°C and supplied with humidified air containing 5% CO2. Since the cell circle is too large to fit in a single image, for each well a horizontal and a vertical overlapping series of images was taken in a crosswise fashion, to generate an accurate picture of the circle dimensions (figure 1b).
Figure 1.
(a) Seeding procedure as described in §2. (b) To determine the size of the cell circle, a series of horizontal and vertical images was taken. (c) Example of cell circle expansion for HBMSC: combined images for the same well at days 0 and 9.
2.3 Image analysis
Customized image analysis software was written in Matlab (v. 7.2, Image Processing Toolbox). For each well, multiple horizontal and vertical images were combined automatically using image correlation (figure 1c). Several contrast enhancement steps were performed, including normalization and filtering with structuring elements to suppress the image background and correct uneven illumination due to condensation. To select the foreground area with the cells, the image background was separated by thresholding. Areas smaller than a critical size were removed. Since the cells in the centre form a continuous sheet, individual cells in the cell layer were identified by selecting the local minima (spots darker than their surroundings, by a certain level) within the foreground area in the original image. The results were inspected visually and a manual correction step was included if necessary.
To obtain the cell density distribution, first the centre of the cell circle was determined. The horizontal position of the centre was defined as the mean position of the cells within the horizontal band of images and analogously for the vertical position. If necessary the centre was corrected manually. For each cell, the radial distance to the centre was computed, after which all cells within regular radial intervals were summed. Cell numbers were normalized by the area of the corresponding circle segment within the photographed area (Matlab bwarea function) to yield the cell density. In this process, a small area along the image boundaries was excluded to remove incomplete cells.
2.4 Proliferation assay
Cell proliferation was assessed independently using the colorimetric WST-1 assay (Roche, UK, Cat. No. 11 644 807 001). To establish the assay kinetics, MG63 cells were added to a 24-well plate at concentrations of 0, 0.066, 0.20, 0.59, 0.89, 1.3, 2.0 and 3.0×105 cells per well (n=3, 1.9 cm2). After 19 h, the medium was replaced with culture medium containing 9% WST-1 reagent and the plate transferred to an incubator. After 0.5, 1, 2, 3, 4 and 6 h, the absorbance was read at 450 nm on an ELx800 microplate reader (BIO-TEK Instruments). Based on these results, a 2 h period was selected for an optimal detection range in subsequent experiments.
MG63 and HBMSCs were added to every well in a 24-well plate at 1×104 cells per well. The HBMSCs used were from the same patient and the experiment started at the same day as the circle assay. The medium was refreshed on a daily basis. Every 24 h for the next 5 days, a column of four wells was harvested, WST-1 medium added and the absorbance read following 2 h of incubation. For each time-point, the measured absorbance was corrected by subtracting the mean of a blank column of wells, without WST-1, in the same plate, which was corrected for the small difference between plain medium and WST-1 medium following 2 h incubation.
2.5 Computational methods
Both the standard (equation (2.1)) and sharp front Fisher equation (equation (2.2)) were evaluated. The standard Fisher's equation reads (Murray 2002; Maini et al. 2004a)
| (2.1) |
The first term in the right-hand side of the equation represents cell migration and the second term cell proliferation. The variable c (cells cm−2) is the cell density, the constant D (cm2 s−1) is the random motility coefficient, comparable to a diffusion coefficient, the constant R (s−1) is the unrestricted growth rate and the constant cmax (cells cm−2) is the maximum cell density at confluence. The same parameters govern the sharp front version of Fisher's equation (Sherratt & Marchant 1996; Murray 2002; Maini et al. 2004a),
| (2.2) |
where the cell mobility term now depends linearly on the cell density.
For the circle migration problem, equations (2.1) and (2.2) were solved in axisymmetric configuration, with a nonlinear finite element method (FEM) code written using Matlab. The method employs standard Galerkin linear elements with Gauss integration. Crank–Nicolson time integration and Newton linearization were used. The FEM code was validated by comparing the numerical results with the analytical solution for a time-dependent problem involving linear diffusion combined with first-order reaction in a cylinder, with a constant non-zero concentration imposed at the boundaries and zero concentration initially (Crank 1975).
The measured initial cell distributions at day 0 were used as initial conditions for the circle migration problems. No-flux boundary conditions apply in the centre and edge of the domain, which was sufficiently large enough to ensure that the edge was never reached by the cell migration front. The problems were solved for the 9-day time period using a 1800 s time-step and a 2.5×10−5 m element size.
For each well, the parameters in equations (2.1) and (2.2), motility coefficient D, proliferation rate R and maximum cell density cmax, were estimated simultaneously based on the summed least-squares criterion for all experimental positions and time-points, using the Matlab fminsearch function with the FEM program in the estimation loop. The same results were obtained for different initial estimates. To provide a global comparison between the different models, a quantitative error measure Eres was defined,
| (2.3) |
where xcomp is the computed value; xexp is the measured value; and Σ represents the total sum over all experimental replicates, time-points and positions. For the WST-1 proliferation assay, an exponential function was used to represent the early phase of proliferation, fitting proliferation rate R and the initial cell density.
2.6 Cellular automaton
To assess the effects of cell size on front propagation in the circle assay, a simple cellular automaton (CA) algorithm was implemented in Matlab, similar to Callaghan et al. (2006). In this approach, the global behaviour of the cell population was modelled based on simple rules for the proliferation of individual cells. Migration was not considered. A rectangular grid was defined in which each position can be occupied by only one cell. The grid spacing h=30 μm was based on the measured maximum MG63 cell density. Initially, the number of cells seeded in the experiment was positioned randomly within the circular seeding area, resulting in a uniform distribution. Cells were updated asynchronously and each time-step the order in which the cells were updated was shuffled randomly. At each time-step, a cell divides with probability Pg or remains idle with probability 1−Pg. In the case of cell division, the daughter cell is placed randomly in one of the surrounding free positions. No division takes place if there is no free position available. The probability Pg was related to the proliferation rate R, as follows: Pg=RΔt, based on the expected increase in the number of cells per time-step. A time-step of 36 s was selected.
3. Results
3.1 Experimental results
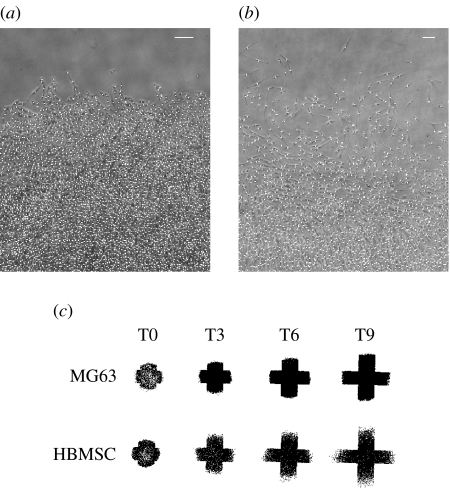
Over the 9-day culture period, the cell circle was noted to increase considerably in size, with an approximate doubling in diameter (figure 1c). Cells in the centre of the circle were densely packed, with MG63 osteosarcoma cells displaying a compact morphology (figure 2a). In contrast, HBMSCs displayed an elongated and spread phenotype (figure 2b, day 9). In these images, the cells identified are indicated by white dots. By plotting the position of all cells identified at different time-points, a global picture of the expanding cell population could be obtained (figure 2c). Marked differences were observed between MG63 and HBMSCs, with the former displaying a relatively sharp front and the latter a more disperse front. This same difference in behaviour was apparent by examination of the cell migration front (figure 2a,b).
Figure 2.
Example of the migration front at day 9 for (a) MG63s and (b) HBMSCs. Positions of cells identified are indicated by white dots. Both scale bars are 200 μm. (c) Overview of cell circle expansion for MG63 and HBMSC (same scale): all identified cell positions plotted for the same well at days 0, 3, 6 and 9.
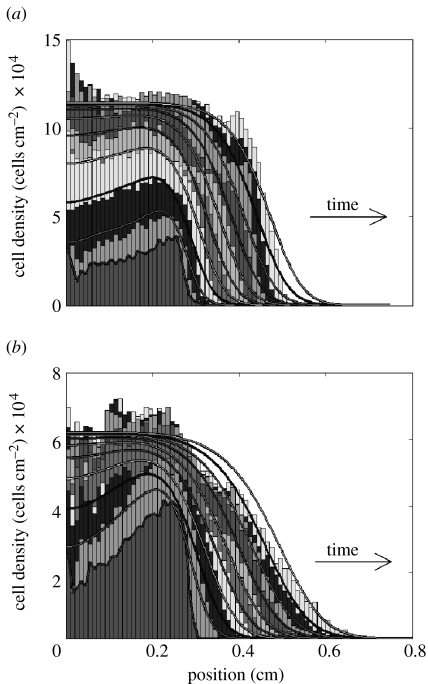
Representative examples of the experimental cell distribution for a single well are shown in figure 3, which shows the cell density as a function of radial distance from the centre of the cell circle. For MG63 cells (figure 3a), a rapid initial increase in cell density in the centre can be observed. While the maximum cell density was being reached in the centre, the cell migration front continued to move outwards at an approximately steady rate. The increase in cell density in the centre appeared to pause between days 3 and 4 (figure 3a). As the cells already appeared confluent at day 3, this represents the start of a phase of denser cell association or possibly even partial overgrowth.
Figure 3.
Comparison of experimental and simulated cell distribution: representative results for the same well. For each day, from day 0 to day 9, the grey histogram bars represent the measured cell density as a function of the radial distance from the centre of the circle for (a) MG63 and (b) HBMSC. The lowest density near the centre is day 0, from there the cell density steadily increased and simultaneously spread outwards to the right, producing a new band every day. The grey lines show the corresponding theoretical model predictions for each day, based on fitting Fisher's equation, equation (2.1).
The more disperse character of HBMSC migration, as seen in figure 2, maps directly to the cell density distribution in figure 3b, displaying a much smoother migration front compared with the sharp front observed for the MG63 cells in figure 3a. The uneven initial distribution, with greater cell presence near the edge of the circle, corresponds to the photomicrograph images and is possibly due to flow as a result of a small amount of fluid leakage from the cylinder. In comparison with the MG63 cells, the maximum cell density for HBMSCs was much lower, at around 50%.
With respect to the seeded cell concentration based on the cylinder diameter, observations from figure 3 show that the initial circle diameter is larger and the cell concentration consequently lower due to the chamfer of the cloning cylinder edge. The estimated total number of cells, based on integrating the density profiles at day 0, is 9.4±0.8×103 (μ±σ) for the HBMSCs and 7.7±0.7×103 for the MG63 cells. These values are in close agreement with the target cell number of 8.7×103 in the seeding suspension based on haemocytometer counting.
3.2 Computational modelling
Figure 3 also illustrates examples of the results of fitting the experimental cell distribution with the standard Fisher model, equation (2.1). The estimated parameter values are given in table 1. For MG63 cells, the initial increase in cell density in the centre of the circle is closely captured (figure 3a), except for the pause between days 3 and 4. However, for this estimated growth rate, the Fisher equation yields a smoother migration front in comparison with the rather sharp front observed in the experiment. Most clearly, this can be seen from the fact that the model predicts a ‘toe-region’, which is absent in the experiment. In Fisher's equation, the migration front velocity depends only on the combination of migration coefficient D and growth rate R. Therefore, it is possible, for the same velocity, to choose a different combination of D and R that yields a different front shape. By increasing the estimated growth rate by a factor of 2, a sharp front shape comparable to the experiment can be obtained. However, this results in a large overprediction of the cell growth rate in the centre.
Table 1.
Estimated migration and proliferation parameters.
| circle assay | MG63 μ±σ (n=5) | σ/μ×100 (%) | HBMSC μ±σ (n=6) | σ/μ×100 (%) |
|---|---|---|---|---|
| Fisher equation (2.1) | ||||
| D (cm2 s−1) | 4.9±0.7×10−9 | 14 | 8.5±2.5×10−9 | 29 |
| R (s−1) | 9.2±0.6×10−6 | 6 | 7.9±1.4×10−6 | 18 |
| cmax (cells cm−2) | 1.2±0.09×105 | 8 | 6.0±0.6×104 | 10 |
| (cm s−1) | 4.2±0.2×10−7 | 4 | 5.1±0.3×10−7 | 6 |
| sharp front Fisher equation (2.2) | ||||
| D (cm2 s−1) | 2.8±0.4×10−8 | 14 | 4.1±1.5×10−8 | 36 |
| R (s−1) | 8.5±0.5×10−6 | 6 | 7.1±1.5×10−6 | 21 |
| cmax (cells cm−2) | 1.2±0.1×105 | 8 | 6.4±1.0×104 | 15 |
| WST-1 assay | MG63 | HBMSC | ||
| R (s−1) | 6.3×10−6 | — | 6.4×10−6 | — |
In contrast to the MG63 cells, the smooth front of the HBMSC population can be closely represented by the standard Fisher model in general (figure 3b). Note that initially between days 0 and 1, compared with the model, the experimental cell distribution appears less diffusive but rather grows in place at a higher rate. As a consequence, the theoretical solution lags behind slightly during the early days of culture. Furthermore, the cell population growing outwards appears to reach a slightly lower maximum density.
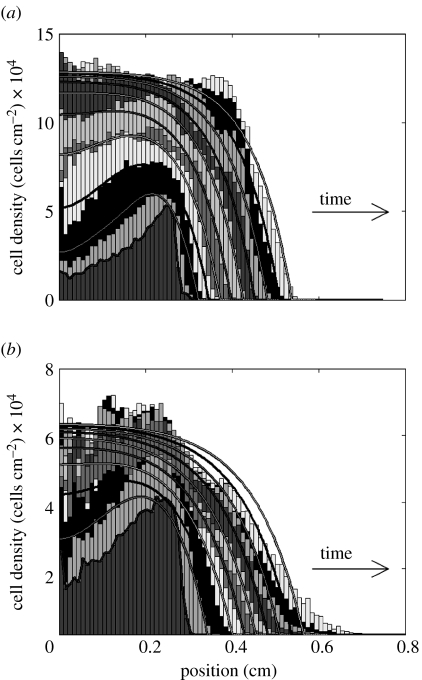
In addition to the standard Fisher equation (equation (2.1)), the sharp front version of Fisher's equation (equation (2.2)) was also evaluated. The type of front solution for this equation does not have a toe-region, but rather rises sharply from zero. For the MG63 cells, figure 4a shows that this sharp front Fisher equation provides a good fit of the experimental cell front, while preserving a correct representation of proliferation in the early phases. Clearly, equation (2.2) cannot represent the front shape for the HBMSCs (figure 4b).
Figure 4.
Comparison of representative experimental and simulated cell distributions in the same well for days 0–9: (a) MG63 and (b) HBMSC. In contrast to the standard Fisher model in figure 3, experimental results were fitted using the sharp front model equation (2.2). The cell distribution is explained in more detail in figure 3.
For the MG63s, the global error measure Eres (equation (2.3)) was 0.11 and 0.10 for the Fisher and sharp front solution fit, respectively. For HBMSCs, the value of Eres was 0.13 for both the Fisher and the sharp front solution. For both cell types, the lack of apparent difference in Eres between the two alternative models results from the fact that Eres is dominated by the error in the interior of the cell circle and is not sensitive enough to variations in front shape.
3.3 Cellular automaton results
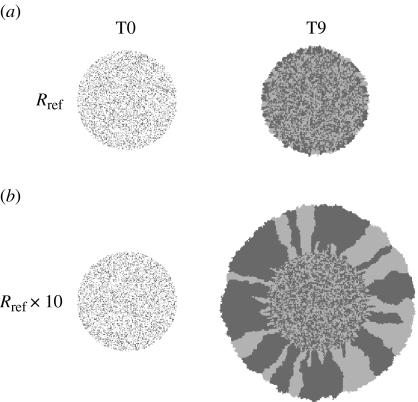
Figure 5a shows the initial and final cell population distribution for the CA model using the growth rate R estimated for MG63s in the circle assay using equation (2.1). Cells were assumed to be immobile. Once the initial distribution has been filled in, this corresponds to growth only at the outer edge. Clearly, it can be seen that this mechanism alone is insufficient to yield the increase in cell circle size observed in the current experiment. Rather, only if the proliferation rate is increased by a factor of 10, a similar increase in size as observed in the experiment can be obtained (figure 5b).
Figure 5.
Cellular automaton results, representative cell distributions at days 0 and 9. For visibility, cells were initially labelled either light or dark grey in equal proportions and subsequently passed this property on to their offspring. (a) Results based on the proliferation rate Rref as estimated for the MG63s using Fisher's equation in the circle assay, but assuming the cells to be immobile. (b) Results for a 10 times higher proliferation rate, without cell movement.
3.4 Proliferation assay results
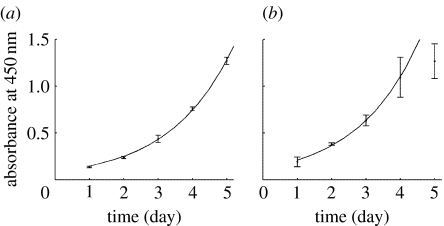
The results of the WST-1 proliferation assay for MG63 and HBMSCs are shown in figure 6. The estimated growth rates R are given in table 1. From figure 6a, the exponential function provides an excellent match of the experimental data for the MG63 cells. However, for the HBMSCs, the variation is larger and only the first four time-points are in close agreement with exponential growth (figure 6b). This departure from exponential behaviour is not surprising since this model can only represent early phase unrestricted growth and does not include effects like crowding. However, there are not enough data to support a logistic model.
Figure 6.
Proliferation kinetics measured by WST-1 assay for (a) MG63s and (b) HBMSCs. To determine the proliferation rate, the experimental results were fitted using a simple exponential function. All data points were used for the MG63s. For the bone marrow cells only the first four points were fitted.
4. Discussion
The experimental system and image analysis method used were able to provide a high resolution picture of cell distributions. These experimental observations could be closely represented using theoretical models based on random cell motion and proliferation. Notably, MG63 and HBMSCs showed distinct behaviour, with the osteosarcoma cell line displaying a sharp migration front and the HBMSCs behaving more dispersedly. The high level of detail with which the cell distributions were mapped enabled a precise assessment of the correspondence between experimental results and theoretical model predictions. This analysis revealed that the standard Fisher equation can closely describe the migration behaviour of the bone marrow cell population, while for the MG63 cells the sharp front model is more appropriate.
4.1 Cellular migration and proliferation parameters
In a similar circle assay, a population of rat marrow stromal osteoblasts on peptide-modified hydrogels expanded at a rate of 1.7–2.7×10−7 cm s−1 (Shin et al. 2004), which is of the same order, although lower, than the front velocity of 4.2–5.1×10−7 cm s−1 (table 1, row 4) found in the present study. Using Fisher's equation, a value of 4.17×10−9 cm2 s−1 has been estimated for the random motility coefficient D of human peritoneal mesothelial cells on plastic substrate, based on the measured front velocity and proliferation rate from the literature (Maini et al. 2004a). This corresponds closely to the value of 4.9±0.7×10−9 cm2 s−1 found using the standard Fisher equation for the MG63s in the present study. Slightly lower values for D ranging between 1.22 and 2.23×10−9 cm2 s−1 have been determined for rat osteoblasts on RGDS and RDGS-modified glass, respectively (Dee et al. 1999). With a value for D of 8.5±2.5×10−9 cm2 s−1, the HBMSCs in the present study displayed a higher mobility compared with both these studies and the MG63s. However, in the literature, up to one order of magnitude higher motility coefficients have been determined in the range of 1.1–6.6×10−8 cm2 s−1 for various cell types and species (Farrell et al. 1990; Sheardown & Cheng 1996; Maini et al. 2004a). Typical parameters for the sharp front model are not known.
Surprisingly, from cell culture experience, the cell proliferation rates determined in the circle assay for the MG63 cell line and HBMSCs were highly similar, with values for R of 9.2±0.6×10−6 and 7.9±1.4×10−6 s−1, respectively. This is higher than the growth rate of 1.18×10−6 s−1 that was found for human mesenchymal stem cells derived from bone marrow, although these cells were expanded in a three-dimensional matrix, which in general can be associated with lower proliferation rates (Zhao et al. 2005). However, the proliferation rates are also higher than the R of 1.9×10−6 s−1 determined for human peritoneal mesothelial cells on plastic (Niedbala et al. 1986). Nevertheless, the high growth rate for the HBMSCs corresponds to a doubling time of 24 h, which is not uncommon for cells in general. It is also important to note that, while representing general cell behaviour, the quantitative results for the HBMSCs were derived for a single patient and thus do not yet provide information on the possible variation for different patients.
4.2 Parameter estimation
By combining computational modelling with cell density measurements, the present circle assay can be started at a low seeding density, in contrast to previous studies that started at monolayer density (Dixit et al. 2001; Dolle et al. 2005). The advantage of using a low initial density, as well as having a detailed picture of the shape of the cell migration front, is that the experiment contains more information. This means that, in contrast to most previous studies, both parameters D and R can be estimated from a single assay.
However, since the migration front propagation depends on a combination of D and R, the uniqueness of the estimated parameters needs to be considered. If the theoretical front velocity v for Fisher's equation is calculated from D and R, it can be seen from table 1 (columns 3 and 5) that the relative variation in v is smaller than that in D and R. (It is important to note at this point that the front velocity for the axisymmetric Fisher equation only approaches a constant value for large radial distance; Murray 2002, p. 444.). Furthermore, the variation in D and R for the HBMSCs (column 5) is much larger than that for the MG63s (column 3). This is mainly due to the relatively small increase in cell density for HBMSCs, while the MG63s show a considerable increase in cell density in the centre (figure 3), which contains much more information for a unique determination of the cell growth rate R than the shape of the migration front alone.
Independent measurement of the proliferation rate R using the WST-1 assay yielded values in the same order as in the circle assay, although 20–30% lower, table 1 (last row). Similar results were obtained for MG63s and HBMSCs. Taking the proliferation rate R from the WST-1 assay and fitting only the motility coefficient D in the circle assay for the MG63s resulted in a poorer fit quality, with too low a rate of cell density increase in the centre and an even smoother migration front as a lower R implies a higher D.
4.3 Image analysis
The image analysis and cell identification method performed very well for the MG63 cells. Phase contrast microscopy provided images with high contrast in which the cells can be clearly distinguished as dark dots surrounded by a lighter area, even in monolayer. Inspection revealed that practically all visible cells were identified. However, while the more peripheral areas can be easily checked for accuracy, cells in the centre become extremely densely packed, so that although the separate dark spots can still be clearly identified, it is hard to judge visually whether cells start to grow partially over each other.
However, for the HBMSCs the method performed less well in general. When compared with MG63, the cells were much more spread out, which provides less contrast and renders the results more sensitive to the image quality and the identification parameters used. Owing to their elongated shape, there is less contrast to distinguish between cells in the centre and as a result of the spindle-like morphology in the periphery double detection of cells can occur. This requires a more careful tuning of the identification parameters depending on the image quality. By varying these parameters, the effects on the resulting cell density distribution have been assessed. It was established that especially the maximum cell density in the centre was sensitive to the minimum cell size and local minima threshold, but that the position of the migration front remains invariant.
Most studies that use daily imaging or time-lapse video for wound-healing assays measure only the migration front position and not the detailed cell distribution (Chan et al. 1989; Maini et al. 2004a; Shin et al. 2004). However, actual cell distributions have been determined by manual counting (Savla et al. 2004; Cai et al. 2007). Other methods include those in which the cells were fixed and stained and the cell distribution was primarily based on the ratio of the local area covered by cells to the total available area (Tranquillo et al. 1988; Chan et al. 1989; Maini et al. 2004a). Laser scanning cytometry has been used to measure the positions of individual cells and determine their density by fixing and staining the nuclei (Haider et al. 2003). However, in the present study, the aim was to determine the evolution of the cell distribution, at both high spatial and high temporal resolution; therefore, instead of fixing the cells, live cells were monitored using the same wells for all time-points. Contrast can be improved in future using live staining techniques, preferably of the nucleus to distinguish cells in monolayer, although staining must not affect cell behaviour during long-term culture (Zahm et al. 1997).
4.4 Model interpretation, proliferation and migration mechanisms in front propagation
In wound-healing studies, it has been observed that depending on the cell type the cell population moves in a unidirectional interconnected way versus a multidirectional separate manner (Chan et al. 1989; Friedl 2004). The question is therefore whether the sharp migration front displayed by the closely packed MG63s really is the result of migration and proliferation or could simply be the result of cell division at the outer edge. To investigate the effect of cell size on front propagation, a CA approach was adopted. The results in figure 5a,b imply that, without mobility, an unrealistic 10 times higher growth rate would be necessary to produce a substantial expansion of the cell circle. This would indicate that mobility is necessary to give the cells space to divide. However, this basic model includes only growth at the edge and does not account for possible effects of cells further within the cell circle dividing and pushing outwards. As opposed to such internal growth, BrdU labelling showed that proliferation was mainly located in a narrow region at the migration front (Simpson et al. 2007). Furthermore, increased proliferation at the wound edge has been found in a wound-healing scratch assay in which cell cycle phase was determined (Haider et al. 2003). In accordance with the present results, it was concluded in that study that migration must play an important role since wound closure was much faster than could be expected based on cell size and doubling time alone. Instead of being pushed, continuous cell sheets can also be pulled by cells at the front, depending on the strength of cell–cell adhesion (Nabeshima et al. 1998; Friedl 2004). For example, by modulating adhesion, ‘cohort type’ versus ‘scattering migration’ can be induced by growth factors such as hepatocyte growth factor/scatter factor (Nabeshima et al. 1998).
With respect to the sharp front model equation (2.2), it has been argued that a diffusion coefficient that increases with cell density implies that cells will move preferably up the gradient (Cai et al. 2006). However, the nonlinear diffusion model describes essentially a population process in which the population will eventually spread out. Nevertheless, it seems unlikely that cells at high density display a higher level of random motion, since the level of hindrance is higher as cells get into each others' way (Abercrombie 1980). In wound-healing assays, the cell motility has indeed been observed to decrease as a function of the distance to the wound edge (Zahm et al. 1997). Furthermore, a decrease in random motility coefficient as a function of cell density has been measured directly by analysing individual cell trajectories in a proliferation assay (Cai et al. 2007). However, on the other hand, introducing a model that incorporates such a decreased random mobility at higher cell concentrations due to hindrance (Cai et al. 2007) would lead to a smoother migration front that cannot match the results for the MG63 case. Furthermore, the toe-region of the migration front, which is determined by D(0), would be the same as in the standard Fisher equation (Simpson et al. 2006).
However, instead of affecting undirected movement, hindrance could result in a directional bias (Abercrombie 1980; Zahm et al. 1997; Cai et al. 2007). For a constant velocity, a shorter persistence time of cell movement, due to a reduced free path length, in the direction of the higher density will lead to a net displacement down the gradient that can be even higher than for the case of unrestricted movement. For example, in the extreme case, a single cell that cannot move left but is free to move right will show a resulting mean displacement to the right, while the average displacement of a single unrestricted cell will remain stationary. Such a direction bias depending on cell density might produce an increasing net cell flux at higher density, yielding a sharper front in accordance with the observed results for the MG63s.
A simple alternative interpretation of equation (2.2) is provided in appendix A (equation (A 4)), where, instead of random motility, it is assumed that cell movement is directed strictly away from the more crowded areas towards increasing available space. Note that in the derivation, the cell velocity depends only on the cell density gradient and not on the absolute density level. Furthermore, this interpretation will be appropriate only if the relative magnitude of the contribution of purely random movement is small when compared with the directional component, since any random movement for cell density approaching zero introduces a diffusion term that leads to a non-sharp front.
Depending on the shape of the migration front also, other nonlinear models can be employed, like, for example, the Nagumo equation (Kenkre & Kuperman 2003). Alternatively, a model has been proposed for chemotactic cellular migration that can represent sharp migration fronts without introducing a nonlinear diffusion term (Landman et al. 2003). Using a CA approach, it was demonstrated how sharp fronts can arise from cell–cell adhesion (Khain & Sander 2006). An interesting, agent-based, computational modelling approach has been applied to wound healing (Walker et al. 2004), which can be used to study the importance of pushing and cell–cell attachment to obtain an improved mechanistic insight.
4.5 Experimental directions
As a starting point for model evaluation, the present study was performed on simple tissue culture plastic. Therefore, as a next step, the approach has to be applied to study the migratory behaviour on surfaces with coatings such as collagen, fibronectin or calcium phosphate, which are of more direct relevance for tissue engineering applications. It is well known that cell migration speed is affected to a large extent by the level of cell–surface interaction. Typically, a bell-shaped curve is to be expected with an optimum cell velocity at intermediate levels of cell–surface interaction, since for low adhesion cells cannot transfer force effectively, while for strong adhesion it becomes difficult to disrupt bonds (Lauffenburger 1989; Dimilla et al. 1991). Furthermore, as the present study was performed under basal medium conditions, it would be interesting to repeat the experiments under osteogenic conditions to see whether a further differentiated state would lead to decreased mobility, which would have important implications for scaffold colonization.
In contrast to the homogeneous MG63 cell line, the cell population isolated from the bone marrow sample is heterogeneous, consisting of different cell types including stem and progenitor cells. Theoretically, it has been shown that just a few highly mobile disperser cells can drive migration at approximately half the speed of what would be the case if the whole population would be highly mobile (Murray 2002, p. 480). It would therefore be interesting to investigate where stem and progenitor cells are located within the cell circle, track how these migrate over time and where their offspring ends up. An experimental approach would be, for example, to separate and label an enriched stem cell population of cells positive for specific antibodies (Letchford et al. 2006) and mix these with a non-labelled fraction negative for these antibodies. This might reveal implications for the type and differentiation state of cells derived by cell expansion from colonies and during scaffold colonization.
5. Summary and perspectives
It can be concluded that a detailed spatiotemporal picture of the evolving cell distribution is required to obtain a more precise quantification and distinguish between the migration characteristics of different types of cell populations. In detailed comparison with such experimental data, assumptions on cellular behaviour can be evaluated by theoretical modelling. In addition, modelling is essential to derive cellular parameters that are independent of the particular assay used. The main advantage of the present approach is that it allowed to distinguish and estimate both the proliferation and the migration parameters in a single assay. This has been done in few other studies (Savla et al. 2004; Cai et al. 2007) and is a recurrent issue in most studies of this type (Dee et al. 1999; Maini et al. 2004a; Shin et al. 2004). By continuing the characterization of relevant cell populations, depending on surface and environmental conditions, it will be possible to make quantitative predictions of cell ingrowth and guide further three-dimensional studies. In this manner, such combined experimental–computational analysis can prove effective in a systematic approach leading to improved control of tissue regeneration and strategies for tissue engineering.
Acknowledgments
We would like to thank Prof. M. Taylor and Dr B. MacArthur for their useful discussions. B.G.S. was supported by a research grant from the BBSRC. Work in Richard Oreffo's laboratory was supported by grants from the BBSRC and EPSRC.
Appendix A.
Conservation of cells is given by
| (A1) |
where c is the cell density; v is the cell velocity; and q is the source term representing cell proliferation. In the standard diffusion model, based on random motion, the averaged velocity from the net cell flux reads
| (A2) |
where D is the random motility coefficient. Alternatively, it can be assumed that the cells do not move randomly, but strictly in a direction away from the more crowded areas. For example, taking the cell velocity proportional to the gradient of the available free space,
| (A3) |
where H (cm2 s−1) is the proportionality constant. Substituting this expression for v in equation (A 1) yields
| (A4) |
which is exactly the same result as equation (2.2) and thus offers an alternative interpretation that does not rely on a higher net cell flux due to an increased undirected mobility at higher cell densities. Such an increased random motility is questionable since a high cell density seems more likely to restrict free movement. Note that in equation (A 4) the proportionality constant was named H to distinguish from the interpretation as the common random migration coefficient D.
References
- Abercrombie M. The croonian lecture, 1978: the crawling movement of metazoan cells. Proc. R. Soc. B. 1980;207:129–147. doi: 10.1098/rspb.1980.0017. [DOI] [Google Scholar]
- Bancroft G.N, Sikavitsas V.I, Van den Dolder J, Sheffield T.L, Ambrose C.G, Jansen J.A, Mikos A.G. Fluid flow increases mineralized matrix deposition in 3D perfusion culture of marrow stromal osteoblasts in a dose-dependent manner. Proc. Natl Acad. Sci. USA. 2002;99:12 600–12 605. doi: 10.1073/pnas.202296599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai A.Q, Landman K.A, Hughes B.D. Modelling directional guidance and motility regulation in cell migration. Bull. Math. Biol. 2006;68:25–52. doi: 10.1007/s11538-005-9028-x. [DOI] [PubMed] [Google Scholar]
- Cai A.Q, Landman K.A, Hughes B.D. Multi-scale modeling of a wound-healing cell migration assay. J. Theor. Biol. 2007;245:576–594. doi: 10.1016/j.jtbi.2006.10.024. [DOI] [PubMed] [Google Scholar]
- Callaghan T, Khain E, Sander L.M, Ziff R.M. A stochastic model for wound healing. J. Stat. Phys. 2006;122:909–924. doi: 10.1007/s10955-006-9022-1. [DOI] [Google Scholar]
- Chan K.Y, Patton D.L, Cosgrove Y.T. Time-lapse videomicroscopic study of in vitro wound closure in rabbit corneal cells. Invest. Ophthmol. Vis. Sci. 1989;30:2488–2498. [PubMed] [Google Scholar]
- Crank J. 2nd edn. Oxford University Press; New York, NY: 1975. The mathematics of diffusion. [Google Scholar]
- Cukierman E, Pankov R, Yamada K.M. Cell interactions with three-dimensional matrices. Curr. Opin. Cell Biol. 2002;14:633–639. doi: 10.1016/S0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- Dee K.C, Andersen T.T, Bizios R. Osteoblast population migration characteristics on substrates modified with immobilized adhesive peptides. Biomaterials. 1999;20:221–227. doi: 10.1016/S0142-9612(98)00161-6. [DOI] [PubMed] [Google Scholar]
- Dimilla P.A, Barbee K, Lauffenburger D.A. Mathematical-model for the effects of adhesion and mechanics on cell-migration speed. Biophys. J. 1991;60:15–37. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit P, Hern-Anderson D, Ranieri J, Schmidt C.E. Vascular graft endothelialization: comparative analysis of canine and human endothelial cell migration on natural biomaterials. J. Biomed. Mater. Res. 2001;56:545–555. doi: 10.1002/1097-4636(20010915)56:4%3C545::AID-JBM1126%3E3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Dolle J.P, Rezvan A, Allen F.D, Lazarovici P, Lelkes P.I. Nerve growth factor-induced migration of endothelial cells. J. Pharmacol. Exp. Ther. 2005;315:1220–1227. doi: 10.1124/jpet.105.093252. [DOI] [PubMed] [Google Scholar]
- El-Ghannam A. Bone reconstruction: from bioceramics to tissue engineering. Expert Rev. Med. Devices. 2005;2:87–101. doi: 10.1586/17434440.2.1.87. [DOI] [PubMed] [Google Scholar]
- Erli H.J, Ruger M, Ragoss C, Jahnen-Dechent W, Hollander D.A, Paar O, von Walter M. The effect of surface modification of a porous TiO2/perlite composite on the ingrowth of bone tissue in vivo. Biomaterials. 2006;27:1270–1276. doi: 10.1016/j.biomaterials.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Farrell B.E, Daniele R.P, Lauffenburger D.A. Quantitative relationships between single-cell and cell-population model parameters for chemosensory migration responses of alveolar macrophages to C5A. Cell Motil. Cytoskel. 1990;16:279–293. doi: 10.1002/cm.970160407. [DOI] [PubMed] [Google Scholar]
- Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr. Opin. Cell Biol. 2004;16:14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Haider A.S, Grabarek J, Eng B, Pedraza P, Ferreri N.R, Balazs E.A, Darzynkiewicz Z. In vitro model of “wound healing” analyzed by laser scanning cytometry: accelerated healing of epithelial cell monolayers in the presence of hyaluronate. Cytometry Part A. 2003;53:1–8. doi: 10.1002/cyto.a.10032. [DOI] [PubMed] [Google Scholar]
- Kenkre V.M, Kuperman M.N. Applicability of the Fisher equation to bacterial population dynamics. Phys. Rev. E. 2003;67:051921. doi: 10.1103/PhysRevE.67.051921. [DOI] [PubMed] [Google Scholar]
- Khain, E. & Sander, L. M. 2006 The role of cell–cell adhesion in wound healing. J. Stat. Phys, Online first (arXiv:q-bio.CB/0610015).
- Krooshoop D.J.E.B, Torensma R, van den Bosch G.J.M, Nelissen J.M.D.T, Figdor C.G, Raymakers R.A.P, Boezeman J.B.M. An automated multi well cell track system to study leukocyte migration. J. Immunol. Methods. 2003;280:89–102. doi: 10.1016/S0022-1759(03)00262-X. [DOI] [PubMed] [Google Scholar]
- Landman K.A, Pettet G.J, Newgreen D.F. Chemotactic cellular migration: Smooth and discontinuous travelling wave solutions. SIAM J. Appl. Math. 2003;63:1666–1681. doi: 10.1137/S0036139902404694. [DOI] [Google Scholar]
- Lauffenburger D. A simple-model for the effects of receptor-mediated cell-substratum adhesion on cell-migration. Chem. Eng. Sci. 1989;44:1903–1914. doi: 10.1016/0009-2509(89)85131-0. [DOI] [Google Scholar]
- Lauffenburger D.A, Horwitz A.F. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/S0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Letchford J, Cardwell A.M, Stewart K, Coogans K.K, Cox J.P, Lee M, Beresford J.N, Perry M.J, Welham M.J. Isolation of C15: a novel antibody generated by phage display against mesenchymal stem cell-enriched fractions of adult human marrow. J. Immunol. Methods. 2006;308:124–137. doi: 10.1016/j.jim.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Maini P.K, McElwain D.L, Leavesley D.I. Traveling wave model to interpret a wound-healing cell migration assay for human peritoneal mesothelial cells. Tissue Eng. 2004a;10:475–482. doi: 10.1089/107632704323061834. [DOI] [PubMed] [Google Scholar]
- Maini P.K, McElwain D.L.S, Leavesley D. Travelling waves in a wound healing assay. Appl. Math. Lett. 2004b;17:575–580. doi: 10.1016/S0893-9659(04)90128-0. [DOI] [Google Scholar]
- Mauney J.R, Volloch V, Kaplan D.L. Role of adult mesenchymal stem cells in bone tissue engineering applications: current status and future prospects. Tissue Eng. 2005;11:787–802. doi: 10.1089/ten.2005.11.787. [DOI] [PubMed] [Google Scholar]
- Mistry A.S, Mikos A.G. Tissue engineering strategies for bone regeneration. Adv. Biochem. Eng. Biotechnol. 2005;94:1–22. doi: 10.1007/b99997. [DOI] [PubMed] [Google Scholar]
- Murray J.D. Mathematical biology. 3rd edn. vol. I. Springer; New York, NY: 2002. [Google Scholar]
- Murray J.D. Mathematical biology. 3rd edn. vol. II. Springer; New York, NY: 2003. [Google Scholar]
- Nabeshima K, Shimao Y, Inoue T, Itoh H, Kataoka H, Koono M. Hepatocyte growth factor scatter factor induces not only scattering but also cohort migration of human colorectal–adenocarcinoma cells. Int. J. Cancer. 1998;78:750–759. doi: 10.1002/(SICI)1097-0215(19981209)78:6%3C750::AID-IJC13%3E3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Niedbala M.J, Crickard K, Bernacki R.J. Adhesion, growth and morphology of human mesothelial cells on extracellular matrix. J. Cell Sci. 1986;85:133–147. doi: 10.1242/jcs.85.1.133. [DOI] [PubMed] [Google Scholar]
- Savla U, Olson L.E, Waters C.M. Mathematical modeling of airway epithelial wound closure during cyclic mechanical strain. J. Appl. Physiol. 2004;96:566–574. doi: 10.1152/japplphysiol.00510.2003. [DOI] [PubMed] [Google Scholar]
- Sheardown H, Cheng Y.L. Mechanisms of corneal epithelial wound healing. Chem. Eng. Sci. 1996;51:4517–4529. doi: 10.1016/0009-2509(96)00299-0. [DOI] [Google Scholar]
- Sherratt J.A, Marchant B.P. Nonsharp travelling wave fronts in the Fisher equation with degenerate nonlinear diffusion. Appl. Math. Lett. 1996;9:33–38. doi: 10.1016/0893-9659(96)00069-9. [DOI] [Google Scholar]
- Shin H, Zygourakis K, Farach-Carson M.C, Yaszemski M.J, Mikos A.G. Attachment, proliferation, and migration of marrow stromal osteoblasts cultured on biomimetic hydrogels modified with an osteopontin-derived peptide. Biomaterials. 2004;25:895–906. doi: 10.1016/S0142-9612(03)00602-1. [DOI] [PubMed] [Google Scholar]
- Simpson M.J, Landman K.A, Hughes B.D, Newgreen F. Looking inside an invasion wave of cells using continuum models: proliferation is the key. J. Theor. Biol. 2006;243:343–360. doi: 10.1016/j.jtbi.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Simpson M.J, Zhang D.C, Mariani M, Landman K.A, Newgreen D.F. Cell proliferation drives neural crest cell invasion of the intestine. Dev. Biol. 2007;302:553–568. doi: 10.1016/j.ydbio.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Takamizawa K, Niu S, Matsuda T. Mathematical simulation of unidirectional tissue formation: in vitro transanastomotic endothelialization model. J. Biomater. Sci. Polym. Edin. 1996;8:323–334. doi: 10.1163/156856296x00336. [DOI] [PubMed] [Google Scholar]
- Tranquillo R.T, Zigmond S.H, Lauffenburger D.A. Measurement of the chemotaxis coefficient for human-neutrophils in the under-agarose migration assay. Cell Motil. Cytoskel. 1988;11:1–15. doi: 10.1002/cm.970110102. [DOI] [PubMed] [Google Scholar]
- Walker D.C, Hill G, Wood S.M, Smallwood R.H, Southgate J. Agent-based computational modeling of wounded epithelial cell monolayers. IEEE Trans. Nanobiosci. 2004;3:153–163. doi: 10.1109/TNB.2004.833680. [DOI] [PubMed] [Google Scholar]
- Zahm J.M, Kaplan H, Herard A.L, Doriot F, Pierrot D, Somelette P, Puchelle E. Cell migration and proliferation during the in vitro wound repair of the respiratory epithelium. Cell Motil. Cytoskel. 1997;37:33–43. doi: 10.1002/(SICI)1097-0169(1997)37:1%3C33::AID-CM4%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Zhao F, Pathi P, Grayson W, Xing Q, Locke B.R, Ma T. Effects of oxygen transport on 3-d human mesenchymal stem cell metabolic activity in perfusion and static cultures: experiments and mathematical model. Biotechnol. Prog. 2005;21:1269–1280. doi: 10.1021/bp0500664. [DOI] [PubMed] [Google Scholar]