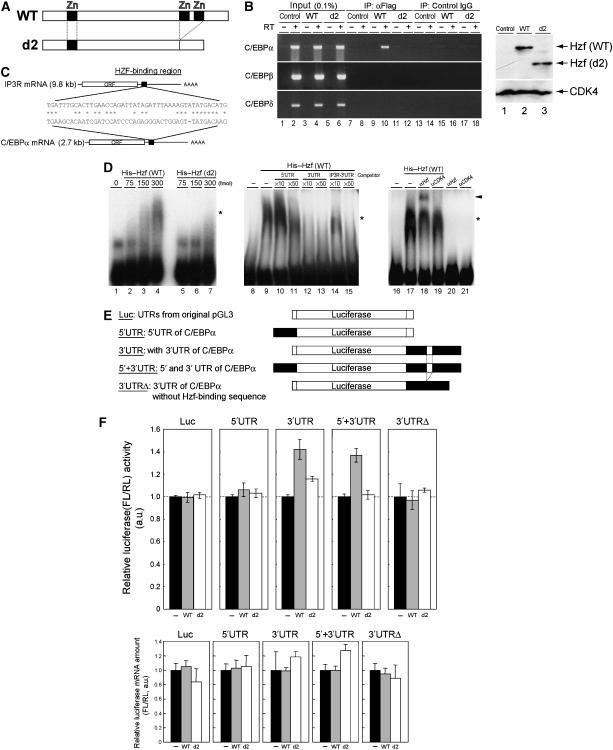
Figure 6.
Hzf associates with C/EBPα mRNA to enhance its translation. (A) Schematic representation of Hzf proteins. (B) 3T3-L1 cells infected with indicated retroviruses were induced to differentiate for 4 days. Following formaldehyde crosslinking, cell lysates were precipitated with antibodies to the Flag epitope or control IgG. RNA eluted from each IP complex was subjected to RT–PCR using C/EBPα, C/EBPβ, or C/EBPδ primers (left panel). The same RT–PCR reaction was performed using 0.1% of supernatant before IP (input). Expression of exogenous Hzf protein was confirmed by immunoblotting with CDK4 as a loading control (right panel). (C) A potential Hzf-binding region in the 3′UTR of C/EBPα mRNA is illustrated. (D) R-EMSA was performed using a radiolabelled RNA probe representing the C/EBPα 3′UTR. The probe was mixed with increasing quantities of bacterially produced wild-type (lanes 2–4) or d2 mutant (lanes 5–7) Hzf proteins. Competition assay was performed on wild-type Hzf proteins with increasing amount of non-radiolabelled C/EBPα 5′UTR (lanes 10 and 11), C/EBPα 3′UTR (lanes 12 and 13), or IP3R 3′UTR (lanes 14 and 15) fragment. Hzf antibody was used for supershift, and CDK4 antibody was used as a control. Complexes were separated by native PAGE and detected by autoradiography. The asterisk and arrowhead indicate Hzf protein–RNA complexes and supershifted complex, respectively. (E) Schematic representation of luciferase reporter mRNAs. Firefly luciferase mRNA in conjunction with 5′ and/or 3′UTRs (either wild-type or mutant sequence that lacks Hzf-binding segment illustrated in (C)) of C/EBPα mRNA was expressed under the regulation of an SV40 promoter and enhancer. (F) Luciferase reporters illustrated in (E) were co-transfected into undifferentiated 3T3-L1 cells together with an SV40-driven Renilla luciferase plasmid used as an internal standard. Upper panel: luciferase activities in each sample were measured, and relative luciferase activities (firefly/Renilla) in each sample were calculated. The relative luciferase activity (FL/RL) of the sample transfected with the control plasmid was normalized to 100% in each group. Lower panel: ratios of firefly and Renilla luciferase mRNA level in each sample were analysed by real-time quantitative PCR analysis.