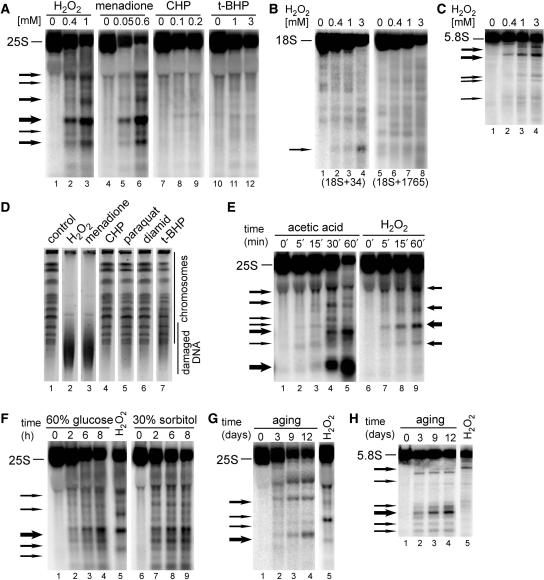
Figure 1.
Ribosomal RNA is fragmented in oxidative stress and apoptotic conditions. Northern hybridizations of total yeast RNA extracted from wild-type W303 cells treated with different stimuli shown above each panel and separated on 1.2% agarose (A, B and E–G) or 6% polyacrylamide (C and H) gels and hybridized with probes against the 25S (007, position +40 relative to the 5′ end), 18S (008, position +34, lanes 1–4 and 029, position +1765, lanes 5–8) or 5.8S rRNA (017, position +30). The position of mature rRNA species is shown on the left of each picture, whereas arrows specify major degradation products. Each experiment was repeated at least 2–3 times, this applies also to Figures 3–7. (A–C) Cells were treated for 200 min with indicated concentrations (mM) of H2O2, menadione, CHP and t-BHP. Hybridizations with probes against 25S (A), 18S (B) and 5.8S (C) are shown. (E) Time courses (min) during exposure to 175 mM acetic acid (lanes 1–5) or 0.6 mM H2O2 (lanes 6–9). (F) Hyperosmotic stress was achieved by addition of 60% glucose (lanes 1–4) or 30% sorbitol (lanes 6–9) for the times indicated (h). RNA from treatment with 0.6 mM H2O2 is shown in lane 5 for comparison. (G–H) Cells were subjected to chronological ageing for times indicated (days, lanes 1–4). RNA from treatment with 0.6 mM H2O2 is shown in lane 5 for comparison. Hybridizations with probes against 25S (G) and 5.8S (H) are shown. (D) Analysis of chromosomal DNA by PFGE. Samples from untreated (lane 1, control) and treated cells for 200 min with 1 mM H2O2 (lane 2), 0.5 mM menadione (lane 3), 0.2 mM CHP (lane 4), 5 mM paraquat (lane 5), 2 mM diamid (lane 6) and 1 mM t-BHP (lane 7) were separated on 1% agarose gels that were stained in ethidium bromide, destained and analysed using Syngene Gene Genius Bioimaging System.