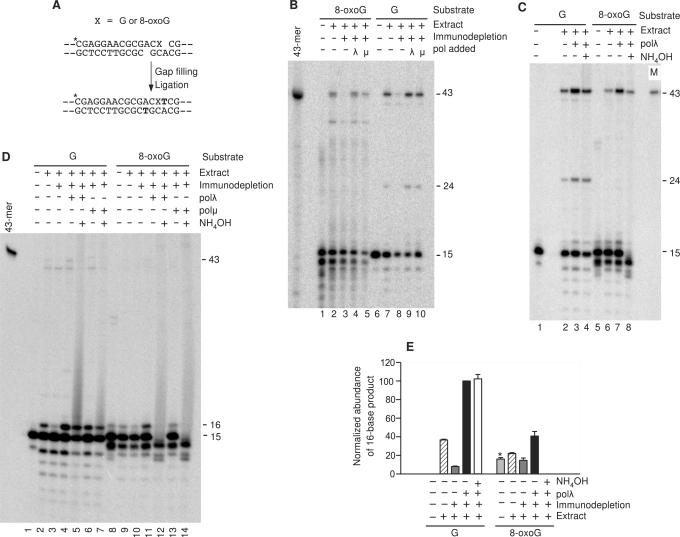
Figure 4.
End joining of a substrate containing an 8-oxoguanine base. (A) Schematic of the substrate and mechanism of formation of the accurate end-joining product (X = guanine or 8-oxoguanine). Bolded T indicates the filled-in base. (B) The substrates were incubated in polλ-depleted or mock-depleted nuclear extracts supplemented with polλ or polμ as indicated, cut with BstXI and AvaI, and analyzed on denaturing gels. The 43-base product corresponds to gap filling and accurate head-to-tail joining of the plasmid, as shown in (A). The 24-base product (formed only with the unmodified substrate) corresponds to accurate head-to-head joining of two plasmid molecules. The band migrating as a ∼14-mer is an apparent spontaneous degradation product of 8-oxoguanine (see text). (C) Same as (B), except that extracts were not immunodepleted, and some samples were treated with NH4OH to cleave 8-oxoguanine residues. The lane marked ‘M’ contains a labeled 43-base marker of the expected sequence. (D) Same as (C), except that the end-joining buffer contained ddTTP instead of dTTP, in order to trap the filled-in but unligated intermediate (16-mer band), and some extracts were immunodepleted as indicated. (E) Quantitative gap-filling data from (D) and a replicate experiment. The abundance of the 16-mer band was calculated for each sample and normalized to the value obtained for the unmodified substrate in the polλ-supplemented extract. The value shown for the 8-oxoguanine substrate (asterisk) includes an apparent degradation product of the initial substrate that comigrates with the 16-mer.