Fig. 5.
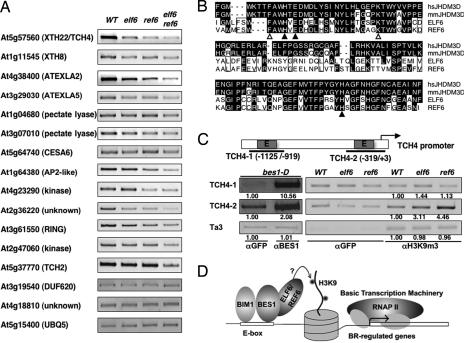
ELF6 and REF6 modulate the expression of some BR-regulated genes, likely by affecting the histone modifications. (A) Many BR-regulated genes were down-regulated in elf6, ref6, and elf6 ref6 double mutants, compared to WT. The genes tested were from BR-induced genes that are reduced in ref6 (Table S1) with three controls. (B) The JmjC domains of ELF6 and REF6 are highly homologous to those from the JHDM3D family from mouse (mm) and human (hs), which demethylate H3K9me3. Key residues required for cofactor binding were conserved [filled triangles, residues for Fe(II) binding and, open triangles, residues for α-ketoglutarate binding]. (C) ChIP assays with bes1-D, WT, elf6, or ref6 with indicated antibodies. The ChIP products were analyzed by PCR with primers from TCH4 gene promoter. αGFP was used as antibody control, and Ta3 served as input control. The numbers under the gel panels indicate relative enrichment of PCR products compared to αGFP control. (D) A model for ELF6 and REF6 function in BES1-mediated gene expression.