Abstract
Mutation of the Caenorhabditis elegans gene unc-89 results in disorganization of muscle A-bands. unc-89 encodes a giant polypeptide (900 kDa) containing two protein kinase domains, PK1 and PK2. Yeast two-hybrid screening using a portion of UNC-89 including PK2, yielded SCPL-1 (small CTD phosphatase-like-1), which contains a C terminal domain (CTD) phosphatase type domain. In addition to the PK2 domain, interaction with SCPL-1 required the putative autoinhibitory sequence, and immunoglobulin (Ig) and fibronectin type 3 (Fn3) domains lying N-terminal of the kinase domain. SCPL-1 also interacts with PK1, and it similarly requires the kinase domain and upstream Fn3 and Ig domains. Analogous regions from the two other giant kinases of C. elegans, twitchin and TTN-1, failed to interact with SCPL-1. The interaction between SCPL-1 and either Ig-Fn3-PK2 or Fn3-Ig-PK1 was confirmed by biochemical methods. The scpl-1b promoter is expressed in the same set of muscles as unc-89. Antibodies to SCPL-1 localize to the M-line and a portion of the I-band. Bacterially expressed SCPL-1 proteins have phosphatase activity in vitro with properties similar to previously characterized members of the CTD phosphatase family. RNA interference knockdown results in a defect in the function of egg-laying muscles. These studies suggest a new role for the CTD phosphatase family, that is, in muscle giant kinase signaling.
INTRODUCTION
The sarcomere is the molecular machine that performs the work of contraction in striated muscle. It is comprised of overlapping, interacting thin and thick filaments and their attachment structures, the M-lines and Z-disks. The assembly and maintenance of the sarcomere involves specific interactions between hundreds of different proteins. Sarcomeres are unusually enriched for very large polypeptides, generally >700,000 Da, including nebulin and titin. These giant proteins are primarily composed of multiple inexact copies of small domains, typically 35–100 residues. From the now extensive knowledge about the many binding partners of the 3-MDa polypeptide titin in vertebrate muscle, several themes have emerged (Lange et al., 2006). First, depending on its location within the titin polypeptide chain, the same type of domain, the immunoglobulin domain, can interact with many different partners. Second, some titin interactors are located only at the sarcomere, whereas others are located at the sarcomere and the nucleus. One of the newest muscle giant modular proteins to be identified is UNC-89 in Caenorhabditis elegans (Ferrara et al., 2005).
The nematode C. elegans is an excellent system in which to study the organization, assembly, and function of striated muscle in a whole organism (Waterston, 1988; Moerman and Fire, 1997; Moerman and Williams, 2006). The gene unc-89 was originally identified in genetic screens for worms that are both slow moving or paralyzed and that have disorganization of their myofilament lattice (Waterston et al., 1980), and then molecularly cloned (Benian et al., 1996). Obscurin was originally identified through a two-hybrid screen by using portions of titin as bait (Bang et al., 2001; Young et al., 2001), and identified as a homologue of UNC-89 (Young et al., 2001). The body wall muscle of unc-89 mutants shows an especial disorganization of the A-band, and for most mutant alleles, lacks M-lines, the structures in the middle of the A-band at which thick filaments are cross-linked (Waterston et al., 1980; Benian et al., 1999). Similarly, obscurin has been shown to have a role in the assembly or maintenance of A-bands: overexpression of a C-terminal portion of obscurin in primary skeletal myotubes results in a disorganization of sarcomeric myosin (Kontrogianni-Konstantopoulos et al., 2004).
unc-89 is a complex gene, which through the use of three different promoters expressed in different sets of muscles, and alternative splicing, generates at least six major protein isoforms UNC-89-A–F (Benian et al., 1996; Small et al., 2004; Ferrara et al., 2005). The largest of these isoforms, UNC-89-B and UNC-89-F, which are each ∼900,000 Da, contain 52 immunoglobulin (Ig) domains; two fibronectin type 3 (Fn3) domains; a triplet of Src homology 3 (SH3), Dbl homology (DH), and pleckstrin homology (PH) domains near their N termini; and two protein kinase domains near their C termini. Two somewhat smaller isoforms, UNC-89-A and UNC-89-E, lack the kinase domains at their C termini. The smallest isoforms, UNC-89-C and UNC-89-D, begin with partial first kinase domains, and each is directed by its own tissue-specific promoter. The human obscurin gene similarly has 3′-most exons encoding two protein kinase domains, with differential splicing producing obscurin A, which lacks the kinase domains (like UNC-89-A and -E); obscurin B (like UNC-89-B and -F), which contains the kinase domains; and two smaller obscurins, one obscurin of which contains intact first and second kinase domains (Bang et al., 2001; Russell et al., 2002; Fukuzawa et al., 2005). Although obscurin contains all the same domains as nematode UNC-89, a difference is that the SH3, DH, PH trio is located near the C terminus rather than near the N terminus as it is in UNC-89. For C. elegans UNC-89, homology modeling indicates that the first kinase domain (PK1) is inactive, but that the second kinase domain (PK2) may have kinase activity (Small et al., 2004). So far, however, neither substrates nor phosphotransferase activity has been demonstrated. Antibodies generated to three distinct regions of UNC-89 localize the proteins exclusively to the M-lines by immunofluorescence microscopy, and more recently, by immuno-electron microscopy (A. Reedy, G. Benian, and P. Hoppe, unpublished data). Vertebrate obscurin is located at the peripheries of both M-lines and Z-disks (Kontrogianni-Konstantopoulos et al., 2003). However, the situation is more complex, with obscurin A (lacking kinase domains) located at the M-line, obscurin B located at the A/I junction and additional isoforms located at the Z-disk and Z/I junction (Bowman et al., 2007).
To help understand how UNC-89 performs its functions, we wanted to identify its binding partners. To begin, we have focused on the C-terminal protein kinase domains. We expected that binding partners of the protein kinase domains would include kinase substrate(s), activator/inhibitor of the kinases or coregulators. Using the yeast two-hybrid method, we found that a novel protein phosphatase binds to each of the protein kinase domains of UNC-89.
MATERIALS AND METHODS
Worm Strains, Culture, and RNA Interference (RNAi)
Nematodes were grown at 20°C on NGM plates with Escherichia coli strain OP50 as food source (Brenner, 1974). Bristol N2 was the wild-type strain, and mutants included unc-89 (su75) (Small et al., 2004), unc-89 (tm752) (Ferrara et al., 2005), and two intragenic deletions from the C. elegans Gene Knockout Consortium, scpl-1(ok1080) and scpl-1(gk283). RNAi for scpl-1 was performed on wild type and unc-89 mutants by using a feeding procedure essentially as described in Kamath and Afhringer (2003). Full-length cDNAs for scpl-1a and scpl-1b were cloned into pPD129.36, and these plasmids or the empty vector were transformed into HT115 (DE3) bacteria before feeding the worms.
Plasmid Construction
For the screening of the two-hybrid library with Ig-Fn3-PK2 kinase, this region of UNC-89 was cloned into the bait plasmid pGBDU-C1 by using BamHI and SalI sites of the vector. This region of UNC-89, called 15/14 in Figure 1A, was polymerase chain reaction (PCR) amplified from the RB2 random primed cDNA library (a gift from Robert Barstead, Oklahoma Medical Research Foundation), by using primers NTSY-15 and NTSY-14 (see Supplemental Table 1 for all primer sequences).
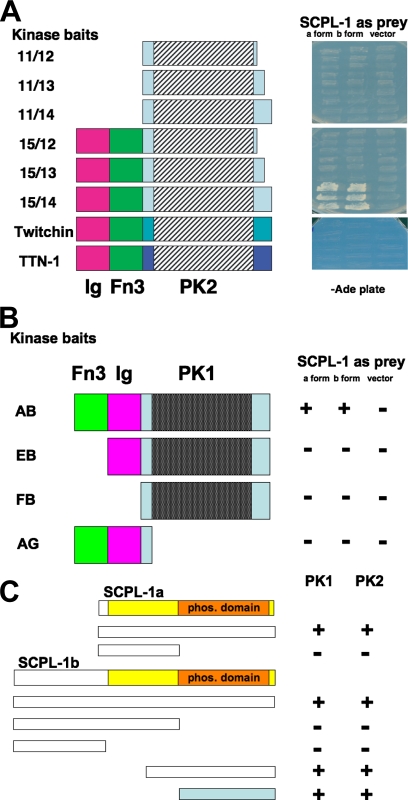
Figure 1.
(A) Yeast two-hybrid assays demonstrate the specificity of the interaction of UNC-89 PK2 with SCPL-1. Left, schematic representation of baits used to test full-length SCPL-1a and -b prey. Right, images of yeast growth on −Ade plates. Note that for UNC-89, interaction requires the catalytic core (PK2) plus the N-terminal Ig and Fn3 domains, and the C-terminal autoinhibitory domain. Note that comparable regions from the two other giant kinases in the worm, twitchin and TTN-1, fail to interact with SCPL-1. (B) The PK1 region of UNC-89 also interacts with SCPL-1 in the yeast two-hybrid assay. Domain mapping indicates that interaction of the PK1 region with SCPL-1 requires, in addition to the catalytic core (PK1), the Fn3 and Ig domains. +, growth and −, no growth on −Ade plates. (C) Only the phosphatase domain of SCPL-1 is required for interaction to the PK regions of UNC-89 in yeast two-hybrid assays. The indicated portions of SCPL-1a and -1b were tested as bait against Fn3-Ig-PK1 (PK1) or Ig-Fn3-PK2 (PK2) prey. +, growth and −, no growth on −Ade plates. The colored bar indicates the minimal region of SCPL-1a/b required for interaction.
To test for interaction between SCPL-1a or -1b prey and various segments of UNC-89 surrounding the PK2 kinase, we first made a series of three baits in the pGBDU-C1 vector, with the same N terminus lying just after the end of the Fn3 domain and varying amounts of sequence C-terminal of the PK2 kinase catalytic core called 11/12, 11/13, and 11/14 (Figure 1A). PCR was used to amplify the corresponding coding sequences from the RB2 cDNA library, by using the same 5′ primer, NTSY-11, with added BamHI site, and three different 3′ primers with added SalI site, NTSY-12, NTSY-13, or NTSY-14. Two additional PK2 region baits were also made similarly, called 15/12 and 15/13 (Figure 1A), by using the primers NTSY-15, -12, and -13.
To construct two-hybrid plasmids expressing various fragments of SCPL-1, PCR-amplified fragments of scpl-1 were cloned into the plasmid pGAD-C1 by using EcoRI and SalI sites of the vector. The primers used for amplification are as follows: SCPL-1-1 and SCPL-1-3 for full length (1-345 amino acids [aa]) of SCPL-1a; SCPL-1-2 and SCPL-1-3 for full length (1-491 aa) of SCPL-1b; SCPL-1-1 and SCPL-1-4 for amino acids 1-156 of SCPL-1a; SCPL-1-2 and SCPL-1-4 for amino acids 1-302 of SCPL-1b; SCPL-1-2 and SCPL-1-5 for amino acids 1-146 of SCPL-1b; and SCPL-1-11 and SCPL-1-3 for amino acids 157-345 of SCPL-1a (amino acids 303-491 of SCPL-1b). The prey plasmid harboring amino acids 245-491 of SCPL-1b was isolated from the library during two-hybrid screening.
To test whether the Ig-Fn3-kinase regions from twitchin and TTN-1 interact with SCPL-1a, -1b,-2, -3a, or -4, these regions of twitchin and TTN-1 were first cloned into the bait plasmid pGBDU-C1 by using the BamHI and SalI sites of the vector. The relevant regions of twitchin were PCR amplified from the RB2 cDNA library by using primers TWI-F and TWI-R, and for TTN-1, the primers were TTN-F and TTN-R.
To determine whether SCPL-1a or -1b preys interact with PK1 region baits, baits called AB, EB, FB, and AG (Figure 1B), were made in pGBDU-C1 by using the SmaI and SalI sites, by insertion of PCR-amplified fragments by using primers U89-PK1-A, U89-PK1-B, U89-PK1-E, U89-PK1-F, and U89-PK1-G.
To test whether UNC-89 Ig-Fn3-PK2 of UNC-89 or comparable regions of twitchin or TTN-1 interact with SCPL-2, -3a, and -4, full-length cDNAs for these other phosphatases were PCR amplified by using the following primers, and then they were inserted into the prey plasmid pGAD-C1 by using BamHI and SalI: for SCPL-2, SCPL-2-F and -R; for SCPL-3a, SCPL-3-F and -R; and for SCPL-4, SCPL-4-F and -R.
To express hemagglutinin (HA)-tagged UNC-89 fragments in yeast, we prepared the following plasmids. The PCR-amplified fragments of the relevant region of UNC-89 PK2 by using the primers U89-PK2-1 and -2 for fragment 11/12; U89-PK2-1 and -3 for fragment 11/13; U89-PK2-1 and -4 for 11/14; U89-PK2-5 and -2 for 15/12; U89-PK2-5 and -3 for 15/13; and U89-PK2-5 and -4 for 15/14 were cloned into pKS-HA8(XbaI) (three HA-tagged vectors) by using the EcoRV site of the vector. The PCR-amplified fragments from the relevant region of UNC-89 PK1 by using the primers as described above were cloned into pKS-HA8(Nhex2) (three HA-tagged vectors) by using EcoRV and SalI sites. HA-tagged UNC-89 11/12, 11/13, 11/14, 15/12, 15/13, and 15/14, AB were cut out with XbaI or NheI from HA-tagged vectors and cloned into pGAP-C-Nhe (yeast expression vector, TRP1 marker) by using the NheI site of the vector.
To express myc-tagged SCPL-1 in yeast, we prepared the following plasmids. The PCR-amplified fragments of full-length SCPL-1a and SCPL-1b by using the primers as described above, were cloned into pKK51 (myc-tagged vector) by using EcoRI and XhoI sites of the vector. Myc-tagged SCPL-1a and myc-tagged SCPL-1b fragments were cut out with SpeI from myc-tagged vector and cloned into pGAPU-C-Nhe (yeast expression vector, URA3 marker) by using the NheI site of the vector.
To express glutathione-S transferase (GST) or maltose-binding protein (MBP) fusions of SCPL-1 in bacteria, we prepared the following. The full-length fragments of SCPL-1a and SCPL-1b were PCR amplified by using primers as described above and cloned into pMAL-KK-1 by using EcoRI and XhoI sites. The fragments of SCPL-1 (1-156 aa of SCPL-1a, 1-302 aa of SCPL-1b, and 1-146 aa of SCPL-1b) were PCR amplified by using the primers described above. Amplified fragments were cloned into pGEX-KK-1 and pMAL-KK-1 by using EcoRI and XhoI sites of the vectors. The plasmids derived from pGEX-KK-1 were used for production of GST fusion proteins, whereas the plasmids derived from pMAL-KK-1 were used for production of MBP fusion proteins.
Yeast Two-Hybrid Screens and Assays
Two-hybrid screening was performed as described in Miller et al. (2006). Use of the two-hybrid method to study protein–protein interaction was performed as described in Mackinnon et al. (2002).
Purification of Bacterially Expressed Proteins
GST or MBP fusions proteins were prepared as described in Mercer et al. (2006).
Assays to Confirm Interaction between SCPL-1 and UNC-89 PK2 or PK1
To confirm an interaction between SCPL-1 and the PK2 region of UNC-89, we expressed in the same yeast cell myc tagged SCPL-1b together with HA-tagged versions of one of the portions of UNC-89 containing PK2 as shown in Figure 1A (11/12, 11/13, 11/14, 15/12, 15/13, 15/14), as described above. Yeast were grown in 50 ml of minimal media plus dextrose, casamino acids, and adenine (SD + CA) to an optical density of ∼0.6 at 600 nm, pelleted, and frozen at −80°C. Each yeast pellet was resuspended in 500 μl of yeast lysis buffer (50 mM HEPES, pH 7.5, 150 mM KCl, 1 mM EGTA, 1 mM EDTA, and complete Mini protease inhibitors, Roche, Indianapolis, IN). The yeast slurries were transferred to 1.5-ml Eppendorf tubes that contained 250 μl of glass beads (425–600 μm in diameter; G-8772, Sigma-Aldrich, St. Louis, MO). Yeast were lysed by vortexing at maximum speed, six bursts of 30 s each, separated by 1 min on ice. After pelleting the beads by centrifugation in a microfuge at 3000 rpm for 5 min, the supernatant was transferred to a fresh tube, and cellular debris was pelleted by centrifugation in a microfuge at 13,200 rpm for 30 min. The resulting supernatant was transferred to a new tube, and its total protein concentration was determined by Bradford assay (Bradford 1976). The HA-tagged PK2 proteins were immunoprecipitated in a total volume of 500 μl for 1 h with shaking at 4°C from 120 μg of protein in immunoprecipitation (IP) buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 0.25% gelatin, complete mini protease inhibitors, and 0.1% NP-40) and 50 μl of a 1:1 slurry of agarose beads conjugated monoclonal antibodies to HA (A2095, Sigma-Aldrich). Beads were pelleted by centrifugation and washed two times with IP buffer and once with IP buffer lacking NP-40. A final hard spin for 5 min was performed, and as much supernatant was aspirated off as possible. Laemmli sample buffer (2×) was added to the pellet, and then the mixture was vortexed 5 s, heated at 95°C for 5 min, vortexed for 20 s, and centrifuged for 5 min to pellet the beads. The supernatant was then carefully transferred to a fresh tube. Two 12% SDS-polyacrylamide gels were run containing either 10 or 40 μl of supernatant, and they were transblotted to nitrocellulose. The blot that had 10 μl of supernatant was reacted with anti-HA (rabbit antibodies, affinity purified, 1:1000; H6908, Sigma-Aldrich), and the blot that had 40 μl of supernatant was reacted with anti-myc (mouse monoclonal purified immunoglobulin, 1:400; M4439, Sigma-Aldrich), reacted with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies, and detected by enhanced chemiluminescence (ECL). Controls included no added yeast lysate, and 10 μg of total protein from any of the yeast to verify the expression of the myc-tagged SCPL-1b.
To confirm an interaction between SCPL-1 and the PK1 region of UNC-89 and to further verify an interaction with the PK2 region of UNC-89, the following procedure was used. Yeast expressing either an HA-tagged PK1 region (segment AB in Figure 1B) or HA-tagged PK2 region (15/14 in Figure 1A) were grown; yeast lysates were prepared; and beginning with 300 μg of total protein, the HA-tagged UNC-89 segments were immunoprecipitated by using anti-HA agarose beads, and washed, as described above. A binding reaction was set up in which the HA-tagged UNC-89 proteins attached to beads were incubated at 4°C for 1 h with shaking in a total volume of 500 μl with either bacterially expressed and purified MBP (10 μg) or MBP-SCPL-1a (10 μg). The beads were washed three times with IP buffer, transferred to clean Eppendorf tubes, centrifuged for 5 min, and as much supernatant was removed as possible. The proteins on the pelleted beads were eluted in Laemmli buffer as described above, and two 10% SDS-PAGE gels were run and transferred to nitrocellulose—one gel contained 5 μl of eluted protein per lane, and the other gel contained 40 μl/lane. The blot that had the 5-μl samples was reacted with anti-HA (described above), followed by anti rabbit Ig-conjugated HRP, and detection by ECL. The blot containing 40-μl samples was incubated with HRP-conjugated anti-MBP (1:5000; New England Biolabs, Beverly, MA) and detected by ECL.
Expression Pattern of scpl-1b Promoter
A 7.5-kb genomic segment that includes the initiator ATG, 5′-untranslated region and putative promoter sequence upstream of scpl-1b was produced by PCR and cloned into pPD95.77 (a gift from A. Fire, Stanford University, Stanford, CA), by using the SphI and BamHI sites. The primers used were SCPL-1b-promo-F and SCPL-1b-promo-R. The promoter-gfp expression plasmid was injected into wild-type strain N2 worms, and one transgenic line was recovered. Green fluorescent protein (GFP) fluorescence images of adults were obtained as described in Qadota et al., 2007.
SCPL-1 Antibodies
For production of anti-SCPL-1b and anti-SCPL-1a, MBP fusion of residues 1-302 of SCPL-1b and MBP fusion of residues 1-156 of SCPL-1a were used as immunogens. For affinity purification of anti-SCPL-1b and anti-SCPL-1a, GST fusion of residues 1-146 of SCPL-1b and GST fusion of residues 1-156 of SCPL-1a were used, respectively. Affinity purification was carried out as described in Mercer et al. (2006).
Western Blot and Immunofluorescence Microscopy
Procedures for preparing worm protein lysates and Western blots were described previously (Mercer et al., 2006). The following worm strains were used to make lysates: wild type (N2), scpl-1(ok1080) and scpl-1(gk283). The blot also contained lysates from yeast expressing myc tagged SCPL-1a and -1b. Yeast lysates were prepared by suspending a yeast pellet from a 2 ml overnight culture in Laemmli buffer and heating at 95° for 5 min. Affinity-purified anti-SCPL-1a was used at 1:200 dilution, and affinity purified anti-SCPL-1b was used at 1:200 dilution. Wild type adult worms were immunostained after fixation by the method described by Nonet et al. (1993). Anti-SCPL-1b was used at 1:100 dilution, and anti-UNC-89 (MH42) was used at 1:200 dilution. Secondary antibodies and confocal microscopy were as described in Qadota et al. (2007).
SCPL-1 Phosphatase Activity Assays
Hydrolysis of phosphate from p-nitrophenyl phosphate (pNPP) was assayed in a 1-ml reaction mixture containing 50 mM Tris acetate, pH 5.0, 10 mM MgCl2, 20 mM pNPP, and MBP or MBP-SCPL-1b (typically 10 μg) as described in Zheng et al. (2005). The amount of protein, the pH, and inclusion of different divalent cations was as described in Zheng et al. (2005), and the use of various phosphatase inhibitors was as described in Kamenski et al. (2004).
Egg-Laying Assays
Examination of drug response to egg laying was performed as described in Trent et al. (1983). Briefly, 96-well plates with 100 μl of M9 containing 5 mg/ml serotonin or 0.75 mg/ml imipramine in each well were prepared. Single adult worms were soaked in each well for 1 h (serotonin) or 1.5 h (imipramine) at room temperature. After incubation, the numbers of laid eggs were counted.
RESULTS
With the goal of identifying binding partners or possible substrates for the protein kinase domains of UNC-89, a portion of the protein containing PK2 was used to screen a yeast two-hybrid library. The bait contained the domains Ig-Fn3-PK2 (Figure 1A, 15/14), including the putative autoinhibitory domain. After screening 8.1 × 106 colonies, 42 positives were confirmed after retransformation, and all of them represented portions of a single gene, B0379.4. As predicted on WormBase and confirmed by sequencing cDNAs, this gene encodes two transcripts, B0379.4a and B0379.4b, by alternative splicing in the 5′ half of the gene. Our prey clones included representatives of both splice isoforms. The proteins encoded by B0379.4a and .4b are 345 and 491 residues, respectively. The only recognizable domain found in these proteins is a protein phosphatase domain (∼170–200 aa) in the C-terminal half of each isoform (Figures 1C and 3A). For reasons explained below, we have renamed B0379.4 as SCPL-1.
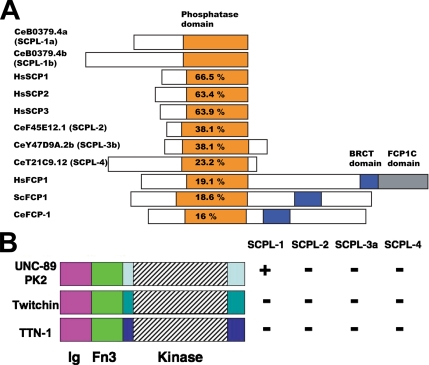
Figure 3.
The SCPL family of proteins in C. elegans and demonstration that UNC-89 PK2 region specifically interacts with SCPL-1. (A) Schematic representation of domain organization of proteins containing CTD type (or FCPH) phosphatase domains. C. elegans has five genes that encode proteins with FCPH domains: one domain called CeFCP-1 is more closely related to FCP proteins in that it has both phosphatase and BRCT domains. The others, including SCPL-1, which interacts with UNC-89, are more closely related to small CTD phosphatases (called SCPs), and they are designated SCP-L. The percentages indicate the percentage of identical amino acids in the ∼200-residue phosphatase domains, compared with the phosphatase in CeB0379.4a (SCPL-1). (B) By yeast two-hybrid assays, the UNC-89 PK2 region interacts with SCPL-1, but not SCPL-2, -3, or -4. The comparable regions of twitchin and TTN-1 fail to interact with any of the SCPL proteins. +, growth and −, no growth on −Ade plates.
To determine which portions of the UNC-89 bait are minimally required to interact with SCPL-1, deletion derivatives of the segment Ig-Fn3-PK2 were tested by two-hybrid against SCPL-1a and -1b full-length prey. As shown in Figure 1A, all the domains are required for interaction, including the C-terminal autoinhibitory region. Because the pattern of domains, Ig-Fn3-kinase, is conserved among the giant protein kinases, we tested the comparable regions from the two other giant kinases in worm muscle, twitchin and TTN-1. As can be seen in Figure 1A, these giant kinases failed to interact with SCPL-1a or -1b.
Given that UNC-89 has a second protein kinase domain, PK1, which is predicted to be catalytically silent (Small et al., 2004), we wondered whether it, too, might interact with SCPL-1. Therefore, fragment Fn3-Ig-PK1 was used as bait to check for interaction with SCPL-1 by two-hybrid analysis. As indicated in Figure 1B, interaction was found, and all the domains, Fn3, Ig, and PK1 need to be present to obtain interaction of the PK1 region with either SCPL-1a or -1b. To determine which portion of SCPL-1a or -1b is required for interaction with either PK1 or PK2 full-length regions, deletion derivatives of SCPL-1a and -1b were tested by two-hybrid analysis. As shown in Figure 1C, the minimal region required is the phosphatase domain.
To provide additional evidence that SCPL-1 interacts with the PK2 and PK1 regions of UNC-89, further experiments were performed. Although each isoform of SCPL-1 could be expressed in bacteria as MBP or GST fusions, segments of UNC-89, either Ig-Fn3-PK2, or Fn3-Ig-PK1 as GST or His-tagged fusions, could not be expressed. In fact, the PK1 or PK2 proteins seemed to significantly reduce the growth rate of E. coli. Therefore, in the same yeast cell, myc-tagged SCPL-1b and individual HA-tagged derivatives of the PK2 region corresponding to those depicted in Figure 1A were expressed. Total protein lysates were prepared and incubated with agarose beads coated with antibodies to the HA tag, washed, and eluted, and portions were run on two gels and blotted. One blot was reacted with anti-HA to detect the presence of the PK2 derivatives. As shown in Figure 2A, appropriately sized proteins were detected from each PK2 derivative. The second blot was reacted with anti-myc to detect the presence of any SCPL-1b that might have been brought down with the PK2 protein. As shown in Figure 2A (bottom), only derivative 15/14 (Ig-Fn3-PK2-autoinhibitory region) was coimmunoprecipitated, consistent with the domain mapping two-hybrid experiment shown in Figure 1A.
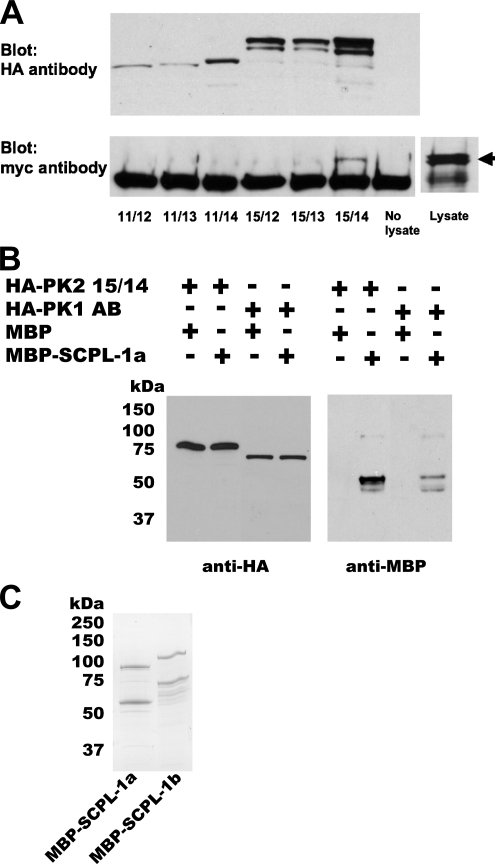
Figure 2.
Verification of interaction between SCPL-1 and UNC-89 PK1 or PK2 regions. (A) In the same yeast cell, myc-tagged SCPL-1b and individual HA-tagged derivatives of the UNC-89 PK2 region were expressed. 11/12, 11/13 … refer to the derivatives presented in Figure 1A. Total protein lysates were incubated with agarose beads coated with antibodies to the HA tag, washed, and eluted, and portions were run on two gels and blotted. One blot was reacted with anti-HA to detect the presence of the PK2 derivatives (top). As shown above, appropriately sized proteins were detected from each PK2 derivative. The other blot was reacted with anti-myc to detect the presence of SCPL-1 that might have been brought down with the PK2 protein (bottom). Only derivative 15/14 (Ig-Fn3-PK2-autoinhibitory region) is coimmunoprecipitated. An arrow designates the position on the blot of myc-SCPL-1b from the lysate. (B) Bacterially expressed MBP-SCPL-1a interacts with yeast expressed HA-PK1 or PK2. Total protein extracts were prepared from yeast expressing either HA-PK2 15/14 (see Figure 1A) or HA-PK1 AB (see Figure 1B), incubated with agarose beads coated with antibodies to HA, washed, incubated with purified, bacterially expressed MBP or MBP-SCPL-1a, and washed. The proteins were eluted, and portions of each sample were run on two gels and blotted. One blot was reacted with anti-HA to detect the presence of the yeast expressed protein, the other blot was reacted with anti-MBP to detect possible binding with MBP or MBP-SCPL-1a. (C) Coomassie stained gel of purified MBP-SCPL-1a and MBP-SCPL-1b. In each case, the top band is likely to be the full-length fusion protein; the second band may have resulted from degradation.
A variation of this experiment was conducted in which yeast was used to express the PK2 or PK1 segments, and E. coli was used to express SCPL-1. Protein extracts were prepared from yeast expressing either HA-PK2 15/14 (Figure 1A) or HA-PK1 AB (Figure 1B), and these UNC-89 segments were immunoprecipitated with anti-HA antibody-conjugated agarose beads. After washing, these beads were incubated with purified, bacterially expressed MBP or MBP-SCPL-1a (Figure 2C), washed, and the proteins were eluted. Portions of each sample were run on two gels and blotted. One blot was reacted with anti-HA to detect the presence of the yeast expressed protein, the other blot was reacted with anti-MBP to detect possible binding with MBP or MBP-SCPL-1a. As shown in Figure 2B, this method demonstrates interaction between MBP-SCPL-1a and either Ig-Fn3-PK2, or Fn3-Ig-PK1. Using MBP-SCPL-1b, we obtained similar results (data not shown).
Application of BLAST and Pfam reveals that the phosphatase domain of B0379.4 (SCPL-1) is most closely related to the protein phosphatase domains of the CTD phosphatase family. The founding member of this family is FCP1 (TFIIF-associating component of CTD phosphatase), which is conserved from yeast to humans, and it is known to dephosphorylate the C-terminal domain (CTD) of the largest subunit of RNA polymerase II. This phosphatase family has no sequence similarity to other phosphatases, except for the motif DXDX(T/V) that is common to both phosphotransferases and phosphohydrolases (Collet et al., 1998). In addition to a phosphatase domain, FCP proteins have a Brca1 C-terminal (BRCT) domain. Other members of this family are generally smaller proteins that lack the BRCT domain, and they are called small CTD phosphatases (SCPs). The function of the SCPs is still being established (see Discussion). Our analysis has revealed that C. elegans has five genes that encode proteins with FCP1 homology (FCPH) or CTD-type phosphatase domains (Figure 3A). One of them is more closely related to FCP (in sequence and the presence of both phosphatase and BRCT domains); thus, it is called CeFCP-1. The others, including B0379.4 (SCPL-1), are more closely related to SCPs; therefore, we call these proteins SCP-like (SCPL). It seems noteworthy that the phosphatase domain of worm SCPL-1 is more closely related to the phosphatase domains of human SCP1, SCP2, and SCP3 (64–67% identical amino acids over a span of ∼200 amino acids), than it is to the other SCPs in C. elegans (ranging from 23 to 38% identity). It is also clear that the phosphatase domain of SCPL-1 is least similar to the phosphatase domains from worm, yeast, and human FCPs (16–19% identity).
To determine whether the PK2 and PK1 regions of UNC-89 interact specifically with SCPL-1, prey plasmids were generated specifying the complete coding sequences for nematode SCPL-2, SCPL-3, and SCPL-4, and they were tested by two-hybrid for interaction. As shown in Figure 3B, when the Ig-Fn3-PK2 segment was tested against all the SCPL proteins, interaction was found only with SCPL-1. In addition, as shown in Figure 3B, when the comparable kinase-containing regions of twitchin and TTN-1 were tested, they failed to interact with any of the SCPL proteins.
Because the two-hybrid library contains cDNAs made from mRNA prepared from whole worms (including all tissues and cell types), it was important to determine whether scpl-1 is expressed in muscle. To do this, we fused in-frame with GFP, 7.5 kb of DNA sequence upstream of the predicted initiator methionine together with codons for a few amino acids of the first exon of scpl-1b. This DNA was used to create transgenic animals, and the sites of GFP expression were recorded. As shown in Figure 4A, scpl-1b is expressed in pharyngeal, vulval, and body wall muscle, the same muscle cells that express unc-89 (Small et al., 2004). This result bolsters the likelihood that SCPL-1 and UNC-89 interact in vivo.
Figure 4.
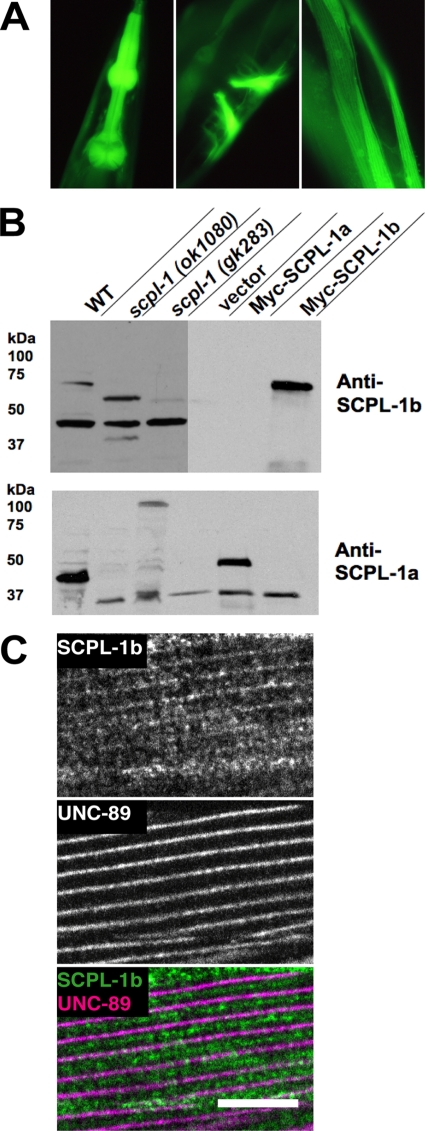
Expression pattern of the SCPL-1b promoter and characterization of antibodies to SCPL-1. (A) SCPL-1b is expressed in the same muscle cells as UNC-89. The exon/intron structure of the SCPL-1b gene is shown, as predicted on WormBase. 7.5 kb of DNA sequence upstream of the initiator methionine together with codons for a few amino acids of the first exon of SCPL-1b were fused in-frame with GFP, and used to create transgenic animals, and the sites of GFP expression were recorded. This promoter is expressed in pharyngeal (left), vulval (center), and body wall muscle (right). (B) Anti-SCPL-1b and anti-SCPL-1a antibodies specifically recognize SCPL-1b and SCPL-1a, respectively. Above is shown immunogens used to generate rabbit antibodies that were later affinity purified using the same regions. Below are Western blots reacted with the designated antibodies. In each case, the left-most three lanes are worm extracts, and the right-most lanes are yeast extracts. WT, wild type; scpl-1(ok1080) and scpl-1(gk283) are intragenic deletions of the scpl-1 gene. Vector, Myc-SCPL-1a, and Myc-SCPL-1b are yeast harboring either the empty vector or myc-tagged SCPL-1 isoforms. (C) Anti-SCPL-1 localize to M-lines and I-bands in body wall muscle. Anti-SCPL-1b and anti-UNC-89 were coincubated with wild-type worms, and the muscle was imaged by immunofluorescence microscopy. The images show a portion of one body wall muscle cell. Weak labeling of the M-line and a portion of the I-band is seen. UNC-89 is a marker of the M-line. Bar, 10 μm.
To determine where the SCPL-1 proteins are expressed and localized, we generated affinity-purified rabbit antibodies that specifically recognize SCPL-1a or SCPL-1b. As shown in Figure 4B, by Western blot, anti-SCPL-1b recognizes yeast-expressed myc-tagged SCPL-1b, but not SCPL-1a, and anti-SCPL-1a recognizes yeast-expressed myc-tagged SCPL-1a but not SCPL-1b. These antibodies recognize the expected sized proteins from C. elegans. Anti-SCPL-1b reacts with an ∼70-kDa protein from wild-type worms likely to be SCPL-1b, and with truncated proteins from the intragenic deletion mutants, ok1080 and gk283. Similarly, anti-SCPL-1a reacts with an ∼50-kDa protein from wild-type worms likely to be SCPL-1a, and this protein is missing from the intragenic deletion mutants.
Anti-SCPL-1b and anti-UNC-89 were coincubated with wild-type worms, and the muscle was imaged by immunofluorescence microscopy. The image presented in Figure 4C shows a portion of one body wall muscle cell. Labeling of the M-line (marked by UNC-89) and a portion of the I-band is seen. In experiments not shown, the I-band location was confirmed by use of known I-band or dense body markers. Similar results were obtained using anti-SCPL-1a (data not shown). Thus, the partial colocalization of SCPL-1 with UNC-89 at the M-line adds further credibility to a functional interaction between SCPL-1 and UNC-89.
Genome-wide RNAi screening (on WormBase) has reported that scpl-1(RNAi) results in a reduction of egg laying or Egl phenotype. We had a difficult time discerning this subtle effect. Therefore, we chose to assay the response to drugs that induce egg laying as described in Trent et al. (1983) and shown in Table 1. Compared with wild type, scpl-1(RNAi) shows a weak response to serotonin, suggesting a defect in function of egg laying muscles. unc-89 mutants showed normal responses to serotonin, unc-89; scpl-1(RNAi) double showed the same defect as scpl-1(RNAi), suggesting that scpl-1 is epistatic to unc-89. Thus, scpl-1 may function downstream of unc-89 in egg laying muscles.
Table 1.
Loss of function of scpl-1 results in a defect in egg laying
| Strain | RNAi | Line | Serotonin |
Imipramine |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| + | (+) | (−) | − | + | (+) | (−) | − | |||
| N2 | scpl-1a full | a | 5 | 3 | 2 | 0 | 8 | 2 | 0 | 0 |
| b | 7 | 1 | 0 | 1 | 9 | 0 | 1 | 0 | ||
| N2 | scpl-1b full | a | 7 | 1 | 1 | 0 | 9 | 1 | 0 | 0 |
| b | 8 | 1 | 1 | 0 | 10 | 0 | 0 | 0 | ||
| N2 | empty vector | a | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 |
| b | 9 | 1 | 0 | 0 | 8 | 2 | 0 | 0 | ||
| unc-89 (tm752) | scpl-1b full | a | 7 | 2 | 1 | 0 | 10 | 0 | 0 | 0 |
| b | 9 | 0 | 1 | 0 | 10 | 0 | 0 | 0 | ||
| unc-89 (tm752) | empty vector | a | 9 | 1 | 0 | 0 | 9 | 1 | 0 | 0 |
| b | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | ||
| unc-89 (su75) | scpl-1b full | a | 8 | 1 | 0 | 1 | 9 | 1 | 0 | 0 |
| b | 7 | 2 | 1 | 0 | 10 | 0 | 0 | 0 | ||
| unc-89 (su75) | empty vector | a | 8 | 2 | 0 | 0 | 10 | 0 | 0 | 0 |
| b | 9 | 1 | 0 | 0 | 10 | 0 | 0 | 0 | ||
+, more than 8 eggs; (+), 4–7 eggs; (−), 1–3 eggs; and −, 0 eggs.
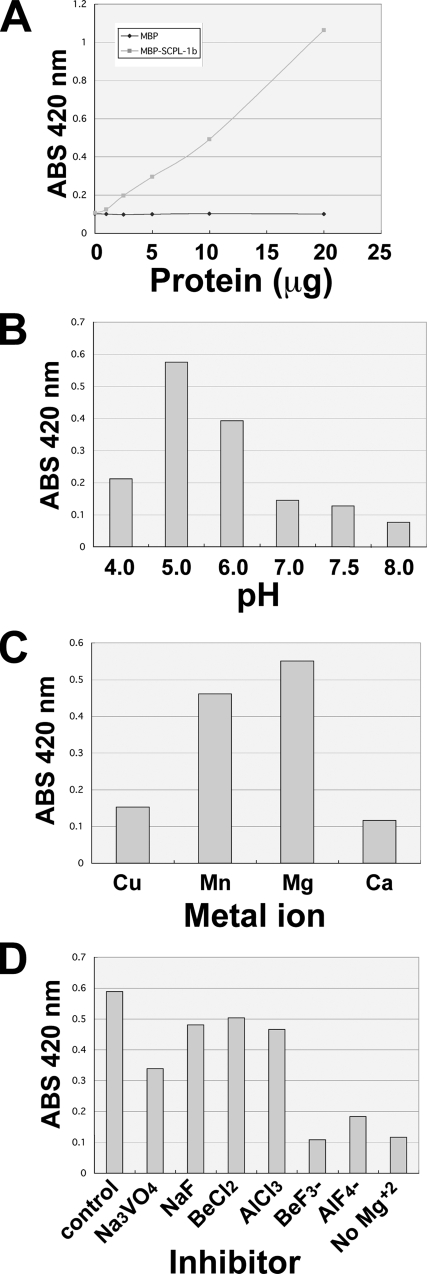
To determine whether SCPL-1 has phosphatase catalytic activity, bacterially expressed MBP fusion proteins of SCPL-1a and SCPL-1b (Figure 2C) were tested in vitro for ability to remove the phosphate from the nonspecific substrate p-nitrophenyl phosphate. Results are shown in Figure 5A for SCPL-1b; similar results were obtained for SCPL-1a; MBP itself showed no activity. As shown in Figure 5A, the amount of product generated was proportional to the amount of added protein. The enzymatic properties are very similar to those of previously characterized FCPs and SCPs (Kamenski et al. (2004); Zheng et al. (2005)), including a pH optimum of ∼5.0, a preference for Mg+2 as divalent cation, and unusual response to small molecule inhibitors. Figure 5D shows that SCPL-1 is not inhibited by the typical phosphatase inhibitors NaF or Na3VO4, but it is inhibited by BeF3− and AlF4.
Figure 5.
Biochemical properties of SCPL-1. SCPL-1b was expressed as an MBP fusion protein in E. coli, and they are shown to have phosphatase activity toward a model substrate, p-nitrophenyl phosphate (A). Enzymatic properties were very similar to previously characterized FCPs and SCPs, including a pH optimum of ∼5.0 (B), preference for Mg+2 (C), and unusual response to small-molecule inhibitors (not inhibited by NaF or Na3VO4 but inhibited by BeF3− and AlF4; shown in D).
DISCUSSION
We have shown that both the putatively catalytically active (PK2) and catalytically inactive (PK1) kinase domains of UNC-89 interact with a novel protein phosphatase, SCPL-1. For UNC-89, this requires the upstream Ig and Fn3 domains, as well as the autoinhibitory region at least in PK2. For SCPL-1, only the phosphatase domain itself is required for interaction with UNC-89. There are two indications of the specificity of this interaction: 1) SCPL-1 does not interact with similar regions of the two other giant titin-like kinases in the worm, twitchin and TTN-1. 2) The UNC-89 PK2 region does not interact with three other related proteins in C. elegans: SCPL-2, -3, and -4, which SAGE data available on WormBase suggest express in body wall muscle. The plausibility of the UNC-89–SCPL-1 interaction is indicated by finding the following: 1) The scpl-1b promoter is expressed in the same sets of muscles (body wall, pharyngeal, and vulval) as unc-89. 2) By antibody staining of body wall striated muscle, SCPL-1 is at least partially colocalized to the M-lines, where it may interact with UNC-89 in situ. Although knockdown of scpl-1 mRNA by RNAi has no obvious phenotype in the structure or function of body wall muscle, it does show a mild defect in egg-laying muscles, as manifest by an abnormal response to serotonin-induced egg laying. unc-89 mutants showed a normal response to serotonin, whereas the unc-89;scpl-1 (RNAi) doubles show the same deficit as spcl-1(RNAi), indicating that scpl-1 is epistatic to unc-89. Therefore, scpl-1 functions downstream of unc-89 in egg-laying muscles. Finally, we have shown that the phosphatase domain of SCPL-1 is an active phosphatase, because phosphatase activity can be demonstrated in vitro with a model substrate. Moreover, because the phosphatase activity is optimal in acidic pH conditions, this might have functional implications. For example, in mammalian muscle, repeated or excessive muscle activity leads to acidosis, and this may stimulate SCPL-1 activity. Alternatively, the phosphatase might be recruited to a different cellular compartment that is acidic (e.g., peroxisomes or autophagosomes).
What might be the biochemical consequences of this interaction? There are at least five possibilities: 1) The UNC-89 putatively active kinase PK2 phosphorylates SCPL-1 to affect its activity. This idea is consistent with SCPL-1 functioning downstream of UNC-89, as suggested by the epistasis results. However, protein kinase assays using yeast-expressed HA-tagged PK2 and bacterially expressed MBP-SCPL-1b were negative (data not shown). 2) The SCPL-1 phosphatase dephosphorylates UNC-89 active kinase PK2 to affect its activity. Precedence for the regulatory affect of phosphorylation on a kinase domain in a giant protein kinase is the tyrosine phosphorylation within the kinase domain of mammalian titin that is one step required in embryonic muscle to relieve autoinhibition (Mayans et al., 1998). 3) Each enzyme, UNC-89 kinase and SCPL-1 phosphatase, has the same substrate, and the two proteins act antagonistically. Vertebrate titin kinase (Grater et al., 2005; Lange et al., 2005), and perhaps the other titin-like kinases such as twitchin, TTN-1, and UNC-89, may be activated (from their autoinhibited states) by small mechanical forces that occur with each contraction/relaxation cycle. Thus, it could be imagined that contraction activates UNC-89 PK2 to phosphorylate the substrate, and during relaxation when UNC-89 PK2 has returned to its autoinhibited state, SCPL-1 phosphatase takes over and dephosphorylates the substrate. 4) The interaction of UNC-89 kinase regions and SCPL-1 forms a structure that recognizes a substrate that neither of the separate enzymes can recognize alone. 5) The interaction of UNC-89 kinase regions with SCPL-1 recruits SCPL-1 to the M-line so that SCPL-1 can dephoshorylate a substrate located there. This scenario does not require that the UNC-89 kinase domains have phosphotransferase activities, but instead act as protein–protein interaction modules. This model is consistent with the finding that both the predicted active PK2 and inactive PK1 bind to SCPL-1. This model of PK1 or PK2 functioning to target SCPL-1 to the M-line is further supported by our finding that prior incubation of PK1 or PK2 with SCPL-1 under conditions that are favorable for binding does not affect the subsequent phosphatase activity of SCPL-1 (data not shown). Further experiments will be required to sort out these different scenarios.
Our studies have pointed to a new function for the CTD phosphatases, that is, in muscle giant protein signaling. The founding member of the CTD phosphatase family is FCP, found in all eukaryotes, and it is known to dephosphorylate serines in the CTD of the largest subunit of RNA polymerase II. This activity of FCP is required for RNA polymerase recycling and global transcription (Yeo et al., 2003). FCP proteins contain both a phosphatase domain (FCPH domain) and a BRCT domain. Higher eukaryotes also contain small CTD phosphatases, proteins possessing the CTD phosphatase domain, but lacking the BRCT domain (Yeo et al., 2003). In vitro, SCPs can dephosphorylate the CTD of RNA polymerase II, but a definitive role as general regulators of transcription has not been demonstrated. So far, there is evidence that SCPs have more specialized functions in transcriptional regulation. For example, SCP1-3 silence neuron-specific gene expression in non-neuronal cells by being recruited by a silencer factor complex (Yeo et al., 2005). Recently, in mammalian cells and Xenopus embryos, SCP1-3, but not FCP1, have been shown to dephosphorylate Smad transcription factors and by doing so, regulate transforming growth factor-β and BMP signaling (Knockaert et al., 2006; Sapkota et al., 2006; Wrighton et al., 2006). Our finding that one of four SCPs in C. elegans specifically interacts with the kinase domains of the giant protein UNC-89 demonstrates yet another function for an SCP protein, a function probably not related to transcriptional regulation. In addition, until our findings, all SCPs have been reported to be nuclear (e.g., Yeo et al., 2003 for SCP1), whereas worm SCPL-1 is located in myofibrils. It is possible that the kinase domains of obscurin (UNC-89 in vertebrates) similarly interact with an SCP protein. It is possible that the true vertebrate orthologue for nematode SCPL-1 has not been identified; for example, although searches of vertebrate proteomes have not revealed an SCP protein with sequence homology outside the phosphatase domain, it is possible that there is a functional homologue of worm SCPL-1 that has an upstream region that forms a similar structure, function, or both. Nevertheless, given the high degree of sequence identity between the ∼200-residue phosphatase domain of worm SCPL-1 to human SCP1, SCP2, and SCP3 (67, 64, and 64%, respectively), even if these phosphatases target different substrate proteins, the recognition sequences that immediately surround the phophorylated serines/threonines might be very similar.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andy Fire for the promoter-less GFP vector, Robert Barstead for the random primed nematode cDNA library, and Kozo Kaibuchi (Nagoya University) for vectors pMAL-KK1 and pKK51. Worm strains were provided by the Caenorhabditis Genetics Center, which is supported by the National Center for Research Resources of the National Institutes of Health. Support for this work was provided by National Institute of Arthritis & Musculoskeletal & Skin Diseases/National Institutes of Health grant AR-051466.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0053) on March 12, 2008.
REFERENCES
- Bang M.-L., et al. The complete gene sequence of titin, expression of an unusual ∼700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Benian G. M., Tinley T. L., Tang X., Borodovsky M. The C. elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian G. M., Ayme-Southgate A., Tinley T. L. The genetics and molecular biology of titin/connectin-like proteins in invertebrates. Rev. Physiol. Biochem. Pharmacol. 1999;138:235–268. doi: 10.1007/BFb0119629. [DOI] [PubMed] [Google Scholar]
- Bowman A. L., Kontrogianni-Konstantopoulos A., Hirsch S. S., Geisler S. B., Gonzalez-Serratos H., Russell M. W., Bloch R. J. Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett. 2007;581:1549–1554. doi: 10.1016/j.febslet.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J. F., Stroobant V., Pirard M., Delpierre G., Van Schaftingen E. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J. Biol. Chem. 1998;273:14107–14112. doi: 10.1074/jbc.273.23.14107. [DOI] [PubMed] [Google Scholar]
- Ferrara T. M., Flaherty D. B., Benian G. M. Titin / connectin-related proteins in C. elegans: a review and new findings. J. Musc. Res. Cell Motil. 2005;26:435–447. doi: 10.1007/s10974-005-9027-4. [DOI] [PubMed] [Google Scholar]
- Fukuzawa A., Idowu S., Gautel M. Complete human gene structure of obscurin: implications for isoform generation by differential splicing. J. Musc. Res. Cell Motil. 2005;26:427–434. doi: 10.1007/s10974-005-9025-6. [DOI] [PubMed] [Google Scholar]
- Grater F., Shen J., Jiang H., Gautel M., Grubmuller H. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J. 2005;88:790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Afhringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kamenski T., Heilmeier S., Meinhart A., Cramer P. Structure and mechanism of RNA polymerase II CTD phosphatase. Mol. Cell. 2004;15:399–407. doi: 10.1016/j.molcel.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Knockaert M., Sapkota G., Alarcon C., Massague J., Brivanlou A. H. Unique players in the BMP pathway: small C-terminal domain phosphatases dephosphorylate smad1 to attenuate BMP signaling. Proc. Natl. Acad. Sci. USA. 2006;103:11940–11945. doi: 10.1073/pnas.0605133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Jones E. M., Van Rossum D. B., Bloch R. J. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol. Biol. Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Catino D. H., Strong J. C., Randall W. R., Bloch R. J. Obscurin regulates the organization of myosin into A bands. Am. J. Physiol. Cell Physiol. 2004;287:C209–C217. doi: 10.1152/ajpcell.00497.2003. [DOI] [PubMed] [Google Scholar]
- Lange S., et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- Lange S., Ehler E., Gautel M. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 2006;16:11–18. doi: 10.1016/j.tcb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Mackinnon A. C., Qadota H., Norman K. R., Moerman D. G., Williams B. D. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr. Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- Mayans O., van der Ven P. F., Wilm M., Mues A., Young P., Furst D. O., Wilmanns M., Gautel M. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- Mercer K. B., Miller R. K., Tinley T. L., Sheth S., Qadota H., Benian G. M. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol. Biol. Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K., Qadota H., Landsverk M. L., Mercer K. B., Epstein H. F., Benian G. M. UNC-98 links an integrin-associated complex to thick filaments in Caenorhabditis elegans muscle. J. Cell Biol. 2006;175:853–859. doi: 10.1083/jcb.200608043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman D. G., Fire A. Muscle: structure, function and development. In: Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R., editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 417–470. [PubMed] [Google Scholar]
- Moerman D. G., Williams B. D. WormBook, editor. Sarcomere assembly in C. elegans muscle. The C. elegans Research Community, WormBook. 2006 doi: 10.1895/wormbook.1.81.1. doi/ 10.1895/wormbook. 1.81.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. L., Grundahl K., Meyer B. J., Rand J. B. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Qadota H., Mercer K. B., Miller R. K., Kaibuchi K., Benian G. M. Two LIM domain proteins and UNC-96 link UNC-97/PINCH to myosin thick filaments in Caenorhabditis elegans muscle. Mol. Biol. Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell M. W., Raeker M. O., Korytkowski K. A., Sonneman K. J. Identification, tissue expression and chromosomal localization of human Obscurin-MLCK, a member of the titin and Dbl families of myosin light chain kinases. Gene. 2002;282:237–246. doi: 10.1016/s0378-1119(01)00795-8. [DOI] [PubMed] [Google Scholar]
- Sapkota G., Knockaert M., Alarcon C., Montalvo E., Brivanlou A. H., Massague J. Dephosphorylation of the linker regions of smad1 and smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-β pathways. J. Biol. Chem. 2006;281:40412–40419. doi: 10.1074/jbc.M610172200. [DOI] [PubMed] [Google Scholar]
- Small T. M., Gernert K. M., Flaherty D. B., Mercer K. B., Borodovsky M., Benian G. M. Three new isoforms of C. elegans UNC-89 containing MLCK-like protein kinase domains. J. Mol. Biol. 2004;342:91–108. doi: 10.1016/j.jmb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Trent C., Tsung N., Horvitz H. R. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics. 1983;104:619–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P., Ehler E., Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J. Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R. H., Thomson J. N., Brenner S. Mutants with altered muscle structure in C. elegans. Dev. Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- Waterston R. H. Muscle. In: Wood W. B., editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 281–335. [Google Scholar]
- Wrighton K. H., Willis D., Long J., Liu F., Lin X., Feng X.-H. Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of smad2 and smad3 to enhance transforming growth factor-β signaling. J. Biol. Chem. 2006;281:38365–38375. doi: 10.1074/jbc.M607246200. [DOI] [PubMed] [Google Scholar]
- Yeo M., Lin P. S., Dahmus M. E., Gill G. N. J. Biol. Chem. 2003;278:26078–26085. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- Yeo M., Lee S. K., Lee B., Ruiz E. C., Pfaff S. L., Gill G. N. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. Science. 2005;307:596–600. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- Zheng H., Ji C., Gu S., Shi B., Wang J., Xie Y., Mao Y. Cloning and characterization of a novel RNA polymerase II C-terminal domain phosphatase. Biochem. Biophys. Res. Commun. 2005;331:1401–1407. doi: 10.1016/j.bbrc.2005.04.065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.