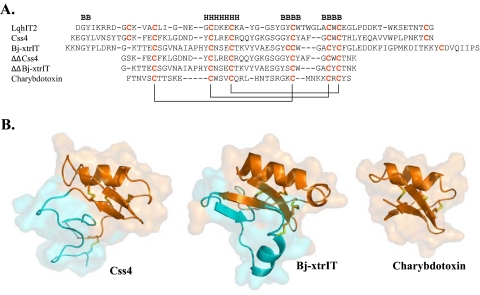
FIGURE 1.
Sequence alignment and three-dimensional structure of representative scorpion toxins. A, sequences were aligned according to the conserved cysteine residues, and the disulfide bonds formed between cysteine pairs are marked in solid lines. Dashes indicate gaps. Secondary structure motifs (B, β-strand; H, α-helix) in Css4 follow the published structure of Cn2 (Protein Data Bank accession 1Cn2). B, schematic diagrams of the Cα model structures of Css4, Bj-xtrIT, and charybdotoxin covered by semitransparent molecular surfaces. The structures of Bj-xtrIT and charybdotoxin are derived from the Protein Data Bank accession numbers 1BCG and 2CRD, respectively. The Css4 model is based on the NMR structure of Cn2 (25) and is spatially aligned with that of Bj-xtrIT and charybdotoxin. The moieties representing ΔΔBj-xtrIT and ΔΔCss4 are colored in orange on the ribbon structure of the parent toxins, and the cyan ribbons represent the deleted N- and C termini. The disulfide bonds are colored in yellow. Panel B was prepared using PyMOL.