Figure 4.
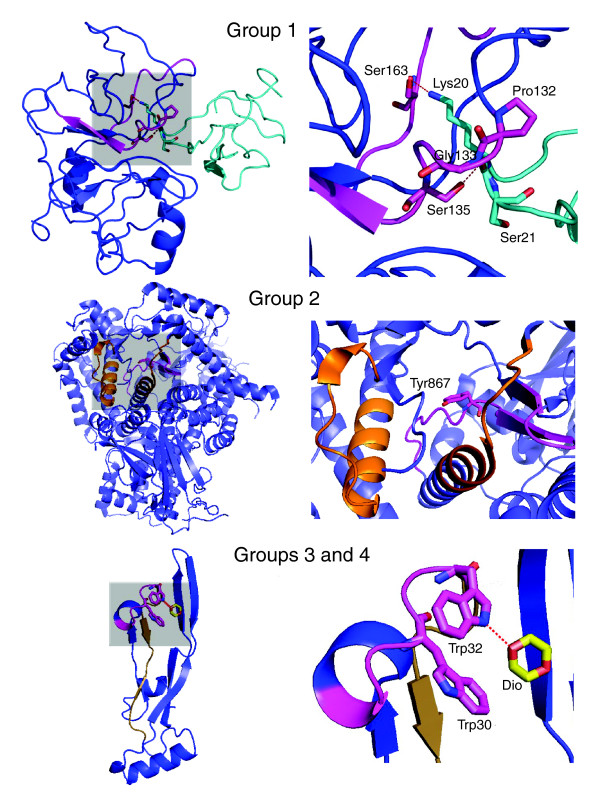
Representative examples from different groups of predictions obtained from our clustering method (see Table 4 for more details). The areas highlighted by gray shading in the left panels are depicted in detail in the right panels. All functionally important regions of the proteins that were identified by our method are shown in magenta with active site/substrate-binding residues in stick representation. Group 1: diagram from PDB entry 1df9 [43], a protease representing examples of fragments for which no PROSITE sequence patterns are available. The residues Pro132 and Gly133 make non-polar interactions with the residues of the NS3 protease (blue) inhibitor (cyan) at P2', while Ser135 and Ser163 make hydrogen bonds to side-chains of Ser21 at P1' and Lys20 at P1, respectively, of the inhibitor. Group 2: diagram from PDB entry 1e7u [44], representing examples for which PROSITE patterns are available but do not overlap with the fragments. The identified functionally relevant region is spatially contiguous to the PROSITE predicted residues; the critical Tyr867 residue implicated in ligand binding is highlighted as a stick model. Groups 3 and 4: diagram from PDB entry 1tgj [45], representing examples where PROSITE pattern overlaps with the fragment. The fragment derived sequence pattern overlaps with the amino-terminal part of the PROSITE pattern (PS00250), which is annotated as a cytokine involved in the repair of tissues. Trp30 and Trp32 interact with the bound dioxane.
