Abstract
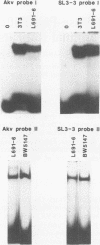
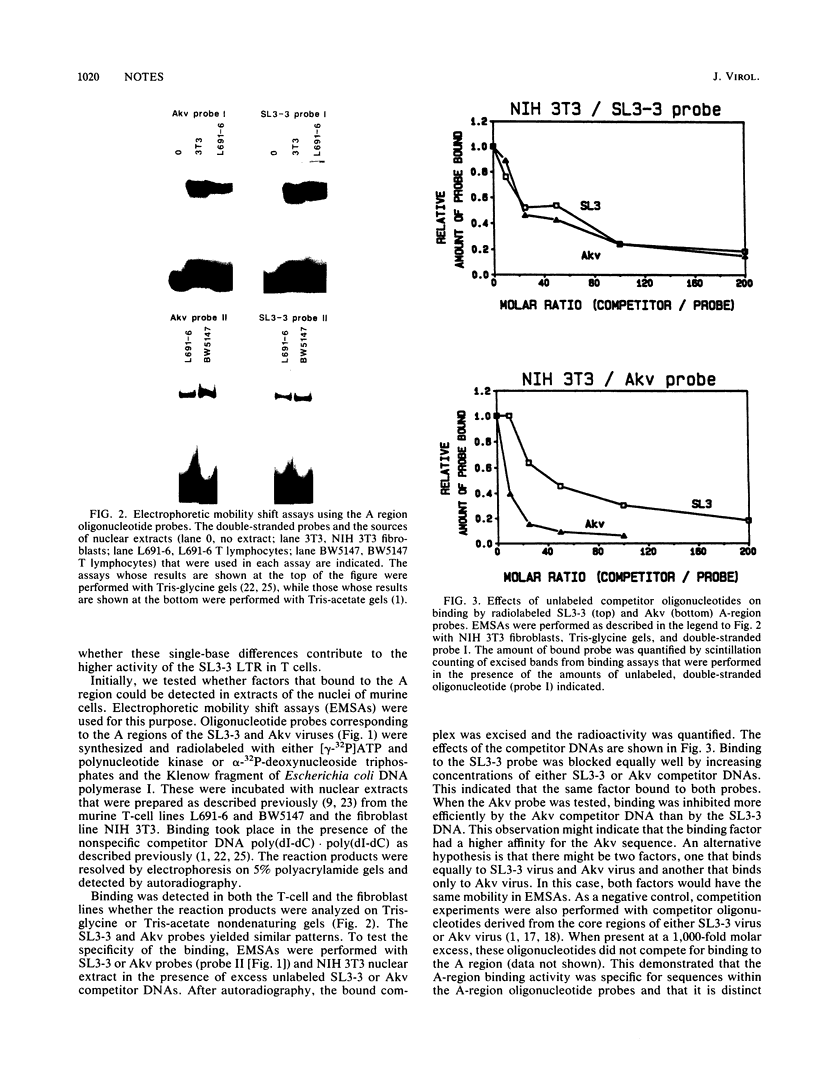
Two single-base-pair differences between the long terminal repeats (LTRs) of the T-lymphomagenic murine retrovirus SL3-3 and nonleukemogenic Akv virus were tested for effects on activity of the LTRs. Evidence was obtained from electrophoretic mobility shift assays for the presence of at least one factor in both T and non-T cells that bound to the region of the viral enhancers that contained the differences. However, no significant differences in activity in expression assays were detected when the two base-pair differences were exchanged between the two LTRs. Therefore, they do not contribute to the higher activity of the SL3-3 LTR in T-lymphoma cell lines.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Puigjaner L. C., Walker J. K., Hall I. H., Birdsall D. L., Ratliff R. L. Wrinkled DNA. Nucleic Acids Res. 1983 Mar 11;11(5):1457–1474. doi: 10.1093/nar/11.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boral A. L., Okenquist S. A., Lenz J. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J Virol. 1989 Jan;63(1):76–84. doi: 10.1128/jvi.63.1.76-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Celander D., Hsu B. L., Haseltine W. A. Regulatory elements within the murine leukemia virus enhancer regions mediate glucocorticoid responsiveness. J Virol. 1988 Apr;62(4):1314–1322. doi: 10.1128/jvi.62.4.1314-1322.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Silver J. E., Frederickson T. N., Hopkins N., Hartley J. W. A 3' end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984 Oct;52(1):248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L., Nicholson J., Hay R. T. Enhancer binding protein (EBP1) makes base and backbone contacts over one complete turn of the DNA double helix. J Mol Biol. 1989 Apr 20;206(4):615–626. doi: 10.1016/0022-2836(89)90570-6. [DOI] [PubMed] [Google Scholar]
- Dai H. Y., Etzerodt M., Baekgaard A. J., Lovmand S., Jørgensen P., Kjeldgaard N. O., Pedersen F. S. Multiple sequence elements in the U3 region of the leukemogenic murine retrovirus SL3-2 contribute to cell-dependent gene expression. Virology. 1990 Apr;175(2):581–585. doi: 10.1016/0042-6822(90)90445-w. [DOI] [PubMed] [Google Scholar]
- DeFranco D., Yamamoto K. R. Two different factors act separately or together to specify functionally distinct activities at a single transcriptional enhancer. Mol Cell Biol. 1986 Apr;6(4):993–1001. doi: 10.1128/mcb.6.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E., Li Y., Fredrickson T. N., Hartley J. W., Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. J Virol. 1989 Jan;63(1):328–337. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorska-Flipot I., Jolicoeur P. DNA-binding proteins that interact with the long terminal repeat of radiation leukemia virus. J Virol. 1990 Apr;64(4):1566–1572. doi: 10.1128/jvi.64.4.1566-1572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg B., Grundström T. Tissue specific sequence motifs in the enhancer of the leukaemogenic mouse retrovirus SL3-3. Nucleic Acids Res. 1988 Jul 11;16(13):5927–5944. doi: 10.1093/nar/16.13.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C. A., Thomas C. Y., Chattopadhyay S. K., Koehne C., O'Donnell P. V. Influence of enhancer sequences on thymotropism and leukemogenicity of mink cell focus-forming viruses. J Virol. 1989 Mar;63(3):1284–1292. doi: 10.1128/jvi.63.3.1284-1292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoSardo J. E., Boral A. L., Lenz J. Relative importance of elements within the SL3-3 virus enhancer for T-cell specificity. J Virol. 1990 Apr;64(4):1756–1763. doi: 10.1128/jvi.64.4.1756-1763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoSardo J. E., Cupelli L. A., Short M. K., Berman J. W., Lenz J. Differences in activities of murine retroviral long terminal repeats in cytotoxic T lymphocytes and T-lymphoma cells. J Virol. 1989 Mar;63(3):1087–1094. doi: 10.1128/jvi.63.3.1087-1094.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovmand S., Kjeldgaard N. O., Jørgensen P., Pedersen F. S. Enhancer functions in U3 of Akv virus: a role for cooperativity of a tandem repeat unit and its flanking DNA sequences. J Virol. 1990 Jul;64(7):3185–3191. doi: 10.1128/jvi.64.7.3185-3191.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley N. R., O'Connell M. A., Sharp P. A., Hopkins N. Nuclear factors that bind to the enhancer region of nondefective Friend murine leukemia virus. J Virol. 1989 Oct;63(10):4210–4223. doi: 10.1128/jvi.63.10.4210-4223.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson P., Hallberg B., Thornell A., Grundström T. Mutant analysis of protein interactions with a nuclear factor I binding site in the SL3-3 virus enhancer. Nucleic Acids Res. 1989 Jun 12;17(11):4061–4075. doi: 10.1093/nar/17.11.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen H. S., Lovmand S., Lovmand J., Jørgensen P., Kjeldgaard N. O., Pedersen F. S. Involvement of nuclear factor I-binding sites in control of Akv virus gene expression. J Virol. 1990 Sep;64(9):4152–4161. doi: 10.1128/jvi.64.9.4152-4161.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro D. J., Sharp P. A., Wahli W. W., Keller M. J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988 Jan-Feb;7(1):47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- Short M. K., Okenquist S. A., Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987 Apr;61(4):1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Golemis E., Fredrickson T. N., Hartley J. W., Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990 Feb;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Hopkins N. Point mutations in the Moloney murine leukemia virus enhancer identify a lymphoid-specific viral core motif and 1,3-phorbol myristate acetate-inducible element. J Virol. 1990 Feb;64(2):543–550. doi: 10.1128/jvi.64.2.543-550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiesen H. J., Bösze Z., Henry L., Charnay P. A DNA element responsible for the different tissue specificities of Friend and Moloney retroviral enhancers. J Virol. 1988 Feb;62(2):614–618. doi: 10.1128/jvi.62.2.614-618.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988 Apr;8(4):1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemur R., Rassart E., DesGroseillers L., Jolicoeur P. Molecular cloning of viral DNA from leukemogenic Gross passage A murine leukemia virus and nucleotide sequence of its long terminal repeat. J Virol. 1983 Feb;45(2):539–546. doi: 10.1128/jvi.45.2.539-546.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F. K., Davison B., Chaffin K. Murine leukemia virus long terminal repeat sequences can enhance gene activity in a cell-type-specific manner. Mol Cell Biol. 1985 Oct;5(10):2832–2835. doi: 10.1128/mcb.5.10.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen P. H., Szurek P. F. The reduced virulence of the thymotropic Moloney murine leukemia virus derivative MoMuLV-TB is mapped to 11 mutations within the U3 region of the long terminal repeat. J Virol. 1989 Feb;63(2):471–480. doi: 10.1128/jvi.63.2.471-480.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]