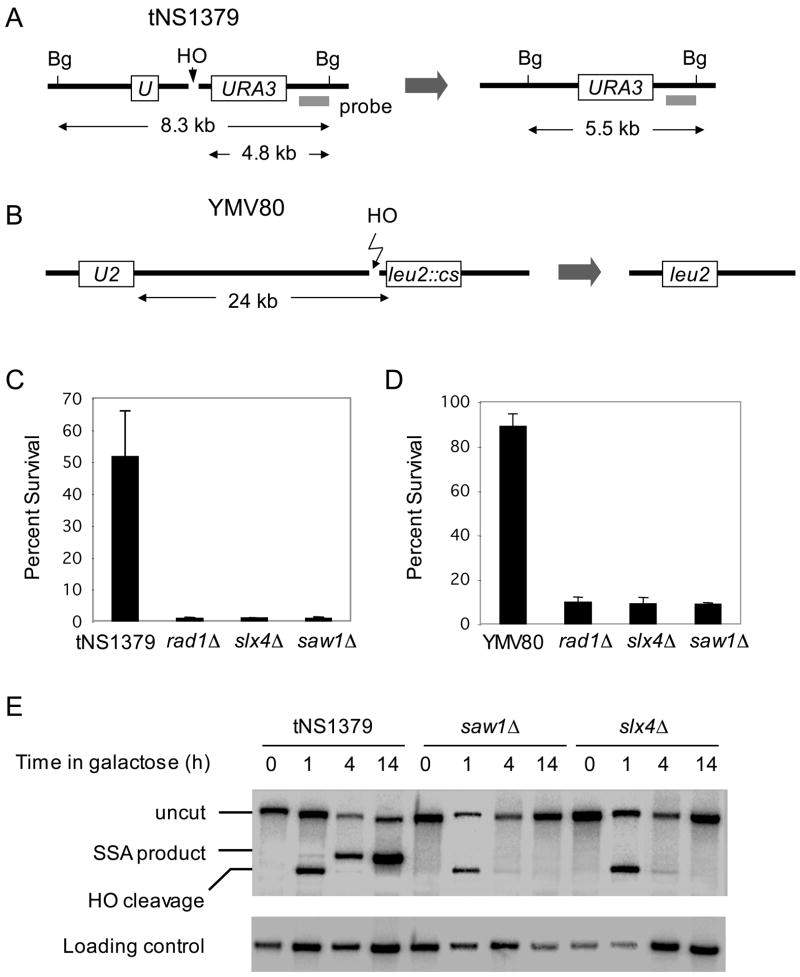
Figure 1. Slx4 and Saw1 are needed for efficient repair of a DSB flanked by direct repeat sequences on a chromosome.
A. SSA between 205 bp ura3 repeats at yeast chromosome V. Survival rate after HO endonuclease induced DSB (C) and Southern blot hybridization of BglII digested genomic DNA (E) with radio-labeled probe next to the URA3 gene to detect SSA product formation are shown for wild type tNS1379 strain and its mutant derivatives. B. SSA between 1.3 kb leu2 repeats that located 24 kb apart on chromosome III. Survival rate of wild type and the SLX4, SAW1, or RAD1 gene deletion derivatives upon HO induction (D) were shown. Data represent means ± standard deviation (s.d.) from three independent experiments.