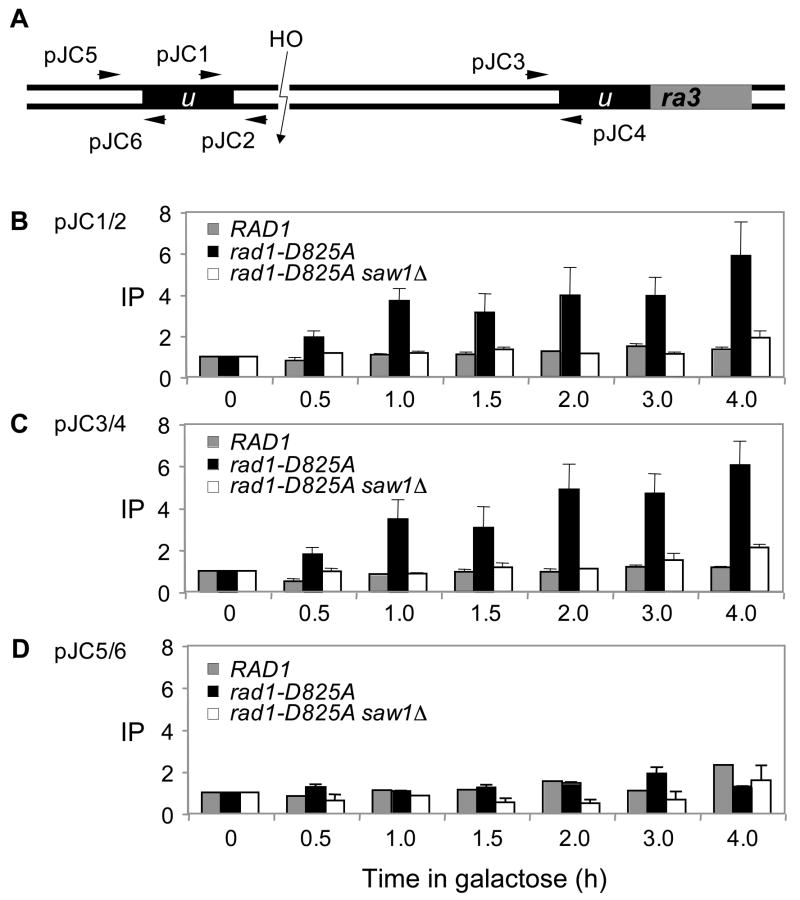
Figure 7. Recruitment of Rad1 at the recombination intermediate carrying 3’-flaps requires Saw1 proteins.
A. HO cleavage and the location of primers (arrows) used in the ChIP assay to detect Rad1 and the nuclease deficient rad1 derivative (rad1-D825A) at the recombination intermediate carrying 3’-flap. The levels of Rad1-3HA or rad1-D825A-3HA in wild type or saw1Δ at the DSB were detected by ChIP assay using an anti-HA antibody. Chromatin was isolated at the indicated time after galactose addition, crosslinked, and fragmented by sonication. After immunoprecipitation and reverse crosslinking, purified DNA was analyzed by qPCR using four sets of primers that anneal 0.2 kb (B, JC1/JC2), 2.4 kb (C, JC3/JC4), and 0.5 kb (D, JC5/JC6) to the DSB, as well as primers specific for the PRE1 gene situated on chromosome V as a control. PCR signals from each primer set at different durations of HO expression were quantified and plotted as a graph. IP represents the ratio of the Rad1 PCR signal before and after HO induction, normalized by the PCR signal of the PRE1 control. Each point is the average of two separate experiments.