Figure 1.
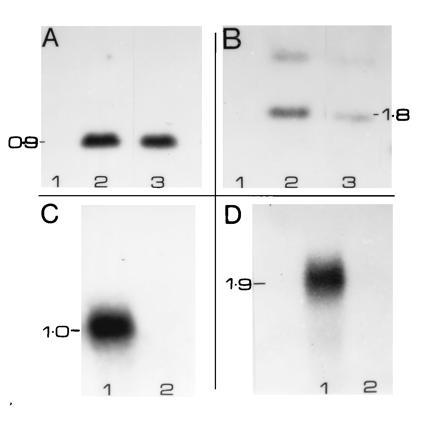
(A) Southern blot analysis of transgenic cotton. Genomic DNA (20 μg each) was isolated from the leaves of DP50 and no. 6888–7 or fibers of no. 7148, and was blotted to nitrocellulose after digestion with XbaI and HindIII restriction enzymes. These enzymes excise the coding region of the phaB gene. Blots were hybridized to 32P-labeled phaB coding region (1 × 108 cpm/μg; 5 × 105 cpm/ml) and washed under stringent conditions (0.1× SSC at 53°C). Autoradiography was done at −70°C for 72 h. Molecular sizes were determined based on standards (1 kb markers; BRL). Lanes: 1, DP50; 2, no. 6888–7; 3, no. 7148. (B) Genomic DNA of no. 6888–7, no. 7148, and DP50 were digested with XbaI and subjected to Southern blot analysis. The blot was hybridized to the coding region of phaC. XbaI digestion is expected to release a 1.8-kb coding region of phaC gene. Lanes: 1, DP50; 2, no. 7148; 3, no. 6888–7. (C) Northern blot analysis of transgenic fiber RNAs. Total RNA (20 μg each) was isolated from 15 DPA fibers of no. 7148 and DP50 control. They were size fractionated on formaldehyde/agarose gels and blotted to nitrocellulose. The blots were hybridized to 32P-labeled (1 × 108 cpm/μg; 0.5 × 105 cpm/ml) insert of phaB and washed under stringent conditions (0.1× SSC at 53°C). Molecular weights were estimated based on 1-kb marker (BRL). Lanes: 1, no. 7148; 2, DP50. (D) Fiber RNAs of no. 7148 and DP50 were hybridized to the insert of the phaC gene. All other conditions were similar to C. Lanes: 1, no. 7148; 2, DP50.
