Abstract
Elucidating the complex combinations of growth factors and signaling molecules that maintain pluripotency or, alternatively, promote the controlled differentiation of human embryonic stem cells (hESCs) has important implications for the fundamental understanding of human development, devising cell replacement therapies, and cancer cell biology. hESCs are commonly grown on irradiated mouse embryonic fibroblasts (MEFs) or in conditioned medium from MEFs. These culture conditions interfere with many experimental conclusions and limit the ability to perform conclusive proteomics studies. The current investigation avoided the use of MEFs or MEF-conditioned medium for hESC culture, allowing global proteomics analysis without these confounding conditions, and elucidated neural cell-specific signaling pathways involved in noggin-induced hESC differentiation. Based on these analyses, we propose the following early markers of hESC neural differentiation: collapsin response mediator proteins 2 and 4 and the nuclear autoantigenic sperm protein as a marker of pluripotent hESCs. We then developed a directed mass spectrometry assay using multiple reaction monitoring (MRM) to identify and quantify these markers and in addition the epidermal ectoderm marker cytokeratin-8. Analysis of global proteomics, quantitative RT-PCR, and MRM data led to testing the isoform interference hypothesis where redundant peptides dilute quantification measurements of homologous proteins. These results show that targeted MRM analysis on non-redundant peptides provides more exact quantification of homologous proteins. This study describes the facile transition from discovery proteomics to targeted MRM analysis and allowed us to identify and verify several potential biomarkers for hESCs during noggin-induced neural and BMP4-induced epidermal ectoderm differentiation.
Human embryonic stem cells (hESCs)1 are perhaps the most promising source of cells for regenerative medicine and treatment of disease. Despite extensive research aimed at elucidating and controlling the processes of self-renewal and lineage-specific differentiation, much remains to be learned regarding the basic cell biology of hESCs before their clinical potential can be realized. Our current knowledge regarding the complex regulation of lineage segregation during development arises primarily from in vivo investigations in invertebrate and mouse. Furthermore manipulation of the signaling pathways initiating and controlling lineage differentiation during development, including the role of bone morphogenic proteins (BMPs), has been expedited by in vitro studies of mouse embryonic stem cells (mESCs). Only recently has considerable research commenced on hESCs.
BMPs were originally characterized based on their ability to induce cartilage and bone formation. They are now known to be multifunctional regulators of development, including many non-osteogenic processes (1). There are as many as 30 different BMP family members classified according to structural similarities. All family members are secreted, undergo dimerization, and initialize signaling pathways by binding cooperatively to their cognate receptors, Type I and Type II BMP receptors (2). These serine/threonine kinase receptors subsequently dimerize and phosphorylate members of the SMAD protein family that translocate to the nucleus where they regulate transcription of target genes (2). BMP signaling can be inhibited in three ways: intracellularly through inhibitory SMADs, at the cognate receptor by pseudoreceptor antagonists, and extracellularly by secreted antagonists. There are seven known extracellular antagonists, including noggin, chordin, follistatin, and sclerostatin. During vertebrate development, the presence of these antagonists, specifically noggin, in the primitive ectoderm inhibits BMP signal transduction, promoting formation of the neural ectoderm, whereas BMP signaling is maintained in the lateral epidermal ectoderm (3). Exposure of mESCs to noggin protein (4) or transfection of noggin expression vectors into mESCs promotes widespread differentiation of primitive neural cells (5). Conversely exposure of mESCs to BMP4 protein antagonizes neural differentiation and has been suggested, with controversy, to either maintain self-renewal or more likely to promote differentiation of different lineages (4).
The conditions that inhibit or promote differentiation of mESCs do not seem to uniformly hold true for hESCs. mESCs are able to retain their undifferentiated state in the presence of leukemia-inhibitory factor (6–9). However, hESCs normally require co-culture with feeder cells, usually mouse embryonic fibroblasts (MEFs), or conditioned medium from these feeder cells to retain their undifferentiated state. As such, the controversy regarding the role of BMP signaling in maintaining hESC pluripotency is most likely due to variable culture conditions and the presence of unknown growth factors during co-culture with MEFs or feeder cell-conditioned medium. However, like mESCs, exposure of hESCs to noggin protein has been widely used to promote differentiation into primitive neural precursors (3, 10–12).
Unfortunately discovery proteomics studies on hESCs have been carried out either on cells grown with feeder cells or grown in the presence of conditioned medium generated from those feeder cells, confounding interpretation of results (13–15). To avoid the potential artifacts introduced by co-culture or exposure to complex mixtures of factors in conditioned medium it is necessary to maintain hESCs free of foreign materials or interactions. Thus, we used defined medium with feeder cell-free conditions to examine lineage-specific differentiation. We expected that BMP4 in feeder-free conditions would either maintain self-renewal by selectively blocking neuronal differentiation or promote differentiation toward an epidermal ectoderm lineage. Additionally we hypothesized that inhibition of the BMP4 signal with noggin would induce neural differentiation. Although these pathways are well characterized qualitatively in mESCs, we aimed at quantitative elucidation with global proteomics technologies through comparisons of control hESCs and hESCs following treatment with noggin and BMP4 separately. These data should then contain the protein markers of pluripotency and early lineage-specific differentiation.
In the current investigation, global quantitative proteomics experiments were completed with iTRAQ labeling combined with two-dimensional (2D) LC TOF/TOF-MS in a bottom-up proteomics approach. This work, including nine experimental replicates (three analytical replicates from each of the three biological replicates; control, +BMP4, and +noggin), led to the identification and quantification of 603 unique proteins. Many proteins implicated in neural cell-specific pathways or epidermal ectoderm differentiation were identified as well as several novel markers of pluripotency and differentiation. After putative protein markers were identified from the global proteomics experiments, a multiple reaction monitoring (MRM) assay was developed. The MRM assay uses two mass filters simultaneously to produce a very precise and sensitive measurement of specific peptides that then act as surrogates for proteins of interest (16). This methodology was utilized to verify protein identifications based on single peptides from the discovery phase global experimentation and to corroborate protein quantification with not only additional peptide identifications but also to obtain quantitative data on specific peptides that distinguish highly homologous protein isoforms. These data show the tremendous capability and facile transition from discovery proteomics to verification of protein targets with MRM and contribute additional information and understanding of both noggin and BMP4 treatment effects on hESCs.
EXPERIMENTAL PROCEDURES
Cell Culture
BG01 hESCs were grown on confluent irradiated MEFs that were plated on tissue culture dishes coated with 0.1% gelatin in growth medium consisting of Dulbecco's modified Eagle's medium/F-12, 20% knock-out serum replacer (KSR), 2 mm glutamine, 0.1% non-essential amino acids, 0.1 mm β-mercaptoethanol, and 4 ng/ml FGF-2. Every 4–8 days, hESCs were manually passaged using Pasteur pipettes (which are pulled over a flame into a thin, flexible strand) as a cutting tool to slice the colonies into small cross-sections. Once dissected, the individual colony pieces were gently lifted from the MEF layer and expanded into culture dishes with fresh feeder cells. Quality control procedures for the hESCs are fully in place, and cells in continuous culture are subjected to regular (quarterly) testing for the following: (i) bacterial and mycoplasma contamination, (ii) a normal karyotype, (iii) maintenance of pluripotency and differentiation capability, and (iv) morphological characteristics including colony size, growth rates, etc.
Differentiation Experiments
BG01 hESCs were manually transferred from MEFs onto 0.1% gelatin-coated dishes, grown for 2 days, visually inspected to ensure removal of MEFs, and then cultured for an additional 7 days in defined medium N2/B27 salts in F-12/Dulbecco's modified Eagle's medium containing 0.2% KSR (Invitrogen) for controls or in the presence of 1 μg/ml noggin protein (R&D Systems, Minneapolis, MN) or 40 ng/ml BMP4 (R&D Systems). Medium was changed daily, and experiments were carried out in triplicate. After 7 days of treatment, BG01 cells from the untreated, noggin and BMP4 treatments were washed and lysed for RNA isolation for qRT-PCR experiments and for whole cellular protein purification for proteomics and Western blotting experiments. Additional replicate cultures were fixed and processed for immunohistochemical localization of pluripotency and lineage-specific proteins as described below.
Immunohistochemistry, Western Blotting, and qRT-PCR
Immunohistochemistry—
After 7 days, hESCs were fixed in 2% paraformaldehyde for 15 min at room temperature and stored in phosphate-buffered saline at 4 °C prior to immunohistochemical localization of cell type-restricted antigens. Cells were permeabilized in 0.1% Triton X-100 for 10 min, blocked to prevent nonspecific antibody binding, and exposed to primary antibody for 2 h at room temperature or overnight at 4 °C. Cells were then washed and exposed to secondary antibody conjugated to a fluorochrome for 30–60 min. Images were captured using a Leitz DM IRB inverted fluorescence microscope and imported into Adobe Photoshop. Antibodies to OCT3/4 (1:100; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and SOX3 (1:1000; from M. Klymkowsky) were localized using corresponding secondary antibodies from Jackson ImmunoResearch Laboratories (West Grove, PA).
Western Blotting—
15 μg of whole cell lysate from each experimental condition, control, and noggin treatment were loaded onto a 4–20% Novex Tris-glycine gel (Invitrogen) under SDS conditions and subjected to electrophoresis for 3 h. The gel-separated proteins were transferred to a nitrocellulose membrane (Invitrogen) and blotted for 2 h at room temperature with the following rabbit primary polyclonal antibodies: α-cytokeratin-8 (1:1000; Abcam ab53708), α-collapsin response mediator protein (CRMP) 4 (1:5000; Abcam ab23951), and the mouse primary monoclonal α-glyceraldehyde-3-phosphate dehydrogenase (1:10,000; Santa Cruz Biotechnology, Inc. sc-58541). Secondary antibodies were goat polyclonal α-rabbit IgG and goat α-mouse IgG conjugated to horseradish peroxidase. Horseradish peroxidase was then detected with ECL Plus (Amersham Biosciences) according to the manufacturer's instructions. Exposed film images were imported into Adobe Illustrator for cropping.
Quantitative RT-PCR—
Reverse transcription was carried out with 1 μg of RNA and random monomers. Quantitative PCR was performed using iQ SYBR Green Supermix and an iCycler (Bio-Rad). Data were analyzed in triplicate using the 2−ΔΔCT method using β-actin as a reference gene for all pairwise comparisons (17).
Proteomics
Sample Preparation for Proteomics—
BG01 hESCs were cultured and treated as described above. Cells were washed, scraped from the dishes, and lysed using Sigma CelLytic M Lysis solution with mammalian protease and phosphatase inhibitor mixtures and 200 mm tris-2-carboxyethylphosphine in 1.5 m Tris-HCl, pH 8.8. The cells were incubated for 15 min at 4 °C, sonicated briefly, and chilled on ice for 10 min. This culture, treatment, and harvesting procedure were repeated a total of three times for each of the three biological replicates. Each solution was cleared by centrifugation for 15 min at 4 °C, and the supernatant was removed and quantified for total protein with a 2D-Quant kit (Applied Biosystems). Aliquots of 100 μg of protein from each of the three treatment conditions and each of the three biological replicates were isolated for proteolytic digestion with sequencing grade trypsin (porcine modified; Promega, Madison, WI) at 1:20 enzyme:protein (w/w) ratio and incubated overnight at 37 °C.
Resulting peptides from each treatment condition were labeled independently with an iTRAQ™ reagent (Applied Biosystems, Foster City, CA) and combined for separation by 2D LC and analysis by MALDI-MS/MS as described previously (18). Briefly each iTRAQ labeling reagent (1 unit in ethanol) was added directly to the protein digest (70% ethanol final), and the mixture was incubated at room temperature for 1 h. The reaction was quenched by addition of 9 volumes 0.1% TFA in water (Optima grade, Fisher Scientific). iTRAQ labeling was completed in the triple-triplex manner; that is, the three different samples, control (labeled with 114.1), BMP4 treatment (labeled with 117.1), and noggin treatment (labeled with 116.1) were labeled separately at the peptide level prior to combination for 2D LC-MALDI-MS/MS. Each of the three biological replicates was analyzed three separate times with 2D LC-MS/MS providing a total of nine replicate experiments. An additional analytical replicate was carried through the cation exchange fractionation step for use in MRM studies.
Chromatography—
Labeled peptides were first separated by strong cation exchange and then by reverse phase liquid chromatography for mass spectrometry analysis on a 4800 MALDI-TOF/TOF instrument. For SCX fractionation, peptides were loaded onto a polysulfoethyl A spin column (SEM HIL-SCX, PolyLC, The Nest Group, Southboro, MA) previously equilibrated with 20% CH3CN in 10 mm KH2PO3 at pH 4.5. For peptide adsorption onto the spin column and for subsequent washing and elution steps, centrifugal force was applied in ∼2-s bursts such that 50 μl of solution passed through the column over a 60-s interval. Loaded peptides were first washed on the column with 800 μl of equilibration buffer and then eluted with 50 μl in a stepwise gradient of increasing salt concentration (35, 50, 65, 80, 95, 115, 135, 155, 180, 205, 350, and 500 mm KCl) in equilibration buffer producing 12 SCX fractions. Eluted peptide fractions were then dried in a vacuum centrifuge and stored at −80 °C until further analysis.
For global proteomics experiments, dried peptide from the 12 SCX fractions were reconstituted with 43 μl of 0.1% TFA in water and separated by reverse phase C18 nano-LC using a 1100 series nano-HPLC instrument equipped with a μWPS autosampler, 2/10 microvalve, MWD UV detector (214 nm), and Micro-FC fraction collector/spotter (Agilent Technologies). With the valve in load position, a 40-μl sample was injected onto an enrichment C18 cartridge (Zorbax 300SB, 5 μm, 5 × 0.3 mm; Agilent Technologies). Mobile phase A, composed of 2.7% CH3CN, 0.1% TFA, was used to desalt the bound peptides at 20 μl/min for 7 min with the effluent directed to waste. Before elution, the enrichment cartridge was placed ahead of an analytical C18 column (Zorbax 300SB, 3.5 μm, 150 × 0.1 mm; Agilent Technologies) previously equilibrated with mobile phase A, and the cartridge was equilibrated with 6.5% mobile phase B, composed of 90% CH3CN, 0.1% TFA. The peptides were then eluted with a linear gradient of 6.5–50% mobile phase B over 65 min at a flow rate of 0.4 μl/min. Column effluent was mixed with MALDI matrix (2 mg/ml α-cyano-4-hydroxycinnamic acid in CH3OH:isopropanol:CH3CN:H2O:acetic acid (12:33.3:52:36:0.7, v/v/v/v) containing 10 mm ammonium phosphate) in a mixing tee (micro Tee, Agilent Technologies). Matrix was delivered with a PHD200 infusion pump (Harvard Apparatus) at 0.8 μl/min. Fractions were spotted at 30-s intervals onto stainless steel MALDI targets (1536 spots/plate; Applied Biosystems).
Mass Spectrometry—
The MALDI target spots were analyzed in the 4800 TOF/TOF™ mass spectrometer (Applied Biosystems/MSD Sciex). First stage MS analysis was completed in positive ion, reflector mode acquiring precursor ions in a mass range of 800–3500 m/z. Tandem MS analysis was completed in a data-dependent manner in which the most abundant 15 peaks were selected per spot with a minimum signal-to-noise ratio of 40. Fragmentation of all peptides was induced by the use of atmosphere as a collision gas with collision energy of 1 kV.
File Conversions—
Peak lists and iTRAQ reporter group area ratios from resulting TOF/TOF tandem mass spectra were extracted by T2Extractor using default parameters (version 2.0). Peak lists were converted to appropriate file types, .mgf and .dta, by the Peak List Conversion Utility (version 2.0) using default parameters (19).
Database Search Algorithms—
Peak lists were searched with SEQUEST (ThermoFinnigan, San Jose, CA; version 27, revision 12), Mascot (Matrix Science, London, UK; version 2.1.0), and X! Tandem (The Global Proteome Machine Organization; version 2006.09.15.1) on a decoy database that contained the forward human IPI sequences concatenated to the reversed human IPI sequences (version 3.16, updated August 2006) (19). The database search used trypsin enzyme specificity, a mass error of 0.5 Da on both parent and fragment ions, a maximum of two missed cleavages, and fixed modifications of methylmethane thiosulfonate on cysteine and iTRAQ reagent on Lys and the peptide N terminus. Variable modifications included deamidation of Asn and Gln; oxidation of Met, His, and Trp; and iTRAQ reagent on Tyr. The decoy database was generated by concatenating the reversed protein sequences to the forward sequences of the entire human IPI database, resulting in a total of 114,732 protein entries. This decoy database strategy allowed for calculation of false positive rates (FPRs) independently for each database search algorithm utilizing the equation FPR = 2 × (FP/(TP + FP)) (20). Peptide identifications were accepted if they were greater than or equal to the 1% FPR calculated by the decoy database for each specific database search engine threshold (shown in Table I). The total number of peptide identifications, both correct and incorrect, were determined and attributed by identification of the forward or reverse sequence, respectively. The number of “acceptable” incorrect peptide identifications can then be calculated from a specified FPR, utilizing the equation above except solving for FP. That is, at a 0.1% FPR, for the SEQUEST results in this data set, one would expect three incorrect peptide identifications. Sorting the data set by decreasing threshold, e.g. ΔCn, and counting the number of forward, i.e. correct hits, until the first three reverse, i.e. incorrect, hits are included, one can report the algorithm threshold-specific ΔCn value that corresponds to that FPR. This is the lowest ΔCn that could be used in a thresholding approach for this data set if one desires an FPR of 0.1%. This was also completed for the Xcorr threshold, but because it is the same data set, one would still expect the 0.1% FPR to still include only three reverse hits. In the same manner, the data set can be sorted by decreasing Xcorr values and counting the number of correct hits until the third incorrect hit aligns the FPR with a Xcorr value of 5.84. This methodology was repeated for Mascot and X!Tandem, but because X!Tandem only has a single threshold score, there is only a single number of estimated correct and incorrect hits for each FPR. Utilizing this methodology, Mascot identifications required ion identity scores ≥40 and ion scores ≥32, SEQUEST identifications required a ΔCn of 0.228 and Xcorr ≥4.75, Mascot identifications required an identity score ≥41.8 and an ion score ≥31.9, and X!Tandem identifications required a log(e) ≥1.89. These thresholds were used for all data as all spectra were only singly charged and represent an FPR of 1%. Protein identifications were accepted only if they contained at least two unique, confidently identified peptides; however, Table II shows the results of the same thresholds with only one confidently identified peptide. Scaffold (version Scaffold-01_06_03, Proteome Software Inc., Portland, OR) was then used to visualize MS/MS-based peptide and protein identifications. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. All individual protein isoforms that the data support equally well are listed in supplemental tables by multiple IPI accession numbers. However, if an individual isoform is listed independently, then at least one distinct, isoform-specific peptide was identified in addition to several redundant peptides.
Table I.
Tabulation of independent thresholds specific to each algorithm and associated number of correct and incorrect peptide identifications found for each calculated FPR
Results were calculated by first determining the expected number of incorrect peptide hits, i.e. those identified reverse sequences for each FPR. That is, at 0.1% FPR we calculated that there will be three reverse, i.e. incorrect, peptide identifications using the equation FPR = 2 × (FP/(TP + FP)) and solving for FP. The integer 2 is used to compensate for the doubling size of the database. All results were sorted by decreasing threshold, e.g. decreasing Xcorr values. The threshold value is then reported that correlates with the expected number of incorrect peptide identifications, e.g. Xcorr of 5.84 for the third false positive representing an FPR of 0.1%. Numbers of correct peptide identifications are those that are equal to or above the threshold cutoff for that FPR.
| SEQUEST
| |||||
|---|---|---|---|---|---|
| Peptide FPR | Incorrect (reverse) peptide sequences | ΔCn | Correct (forward) peptide sequences | Xcorr | Correct (forward) peptide sequences |
| % | |||||
| 0.10 | 3 | 0.4 | 366 | 5.84 | 115 |
| 0.50 | 17 | 0.262 | 1371 | 5.13 | 287 |
| 1 | 33 | 0.228 | 1709 | 4.75 | 447 |
| 5 | 167 | 0.149 | 2483 | 3.94 | 1134 |
| Mascot
| |||||
|---|---|---|---|---|---|
| Peptide FPR | Incorrect (reverse) peptide sequences | Identity score | Correct (forward) peptide sequences | Ion score | Correct (forward) peptide sequences |
| % | |||||
| 0.10 | 2 | 42.9 | 1099 | 46.4 | 1324 |
| 0.50 | 9 | 42 | 1990 | 35.1 | 2360 |
| 1 | 18 | 41.8 | 2236 | 31.9 | 2761 |
| 5 | 90 | 38.1 | 3541 | 24.5 | 3541 |
| X!Tandem
| |||
|---|---|---|---|
| Peptide FPR | Incorrect (reverse) peptide sequences | Log(e) score | Correct (forward) peptide sequences |
| % | |||
| 0.10 | 2 | 3.28 | 2167 |
| 0.50 | 10 | 2.28 | 2919 |
| 1 | 20 | 1.89 | 3285 |
| 5 | 99 | 1.33 | 3901 |
Table II.
Comparison of number of proteins identified by each search algorithm with decoy database FPR calculations and PeptideProphet and ProteinProphet probability calculations for both a minimum of one and two peptides per protein
For each FPR, all protein identifications were filtered by algorithm-specific peptide threshold cutoffs: SEQUEST: Xcorr and ΔCn; Mascot: identity and ion scores; and X!Tandem: log(e) value. PP, PeptideProphet & ProteinProphet probability.
| Algorithm | No. of proteins
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1% FPR, 2 peptides | 1% FPR, 1 peptide | 5% FPR, 2 peptides | 5% FPR, 1 peptide | 95% PP, 2 peptides | 95% PP, 1 peptide | 90% PP, 2 peptides | 90% PP, 1 peptide | |
| SEQUEST | 577 | 1176 | 631 | 1301 | 574 | 726 | 632 | 765 |
| Mascot | 587 | 1107 | 636 | 1159 | 578 | 713 | 630 | 720 |
| X!Tandem | 596 | 1256 | 648 | 1407 | 585 | 720 | 641 | 733 |
Quantification of Peptides and Proteins—
Confidently identified peptides and proteins from global experimentation passing the 1% FPR for each database search algorithm were analyzed for relative quantification by an in-house developed software program described previously (18). Briefly normalization of raw peak areas was accomplished by matching the quantiles of the distributions of each treatment iTRAQ reporter group (116.1 and 117.1) to the quantiles of the control iTRAQ reporter group (114.1). To determine relative ratios, consideration was given that each protein can potentially be identified by a number of different peptides and that every unique peptide can be measured multiple times. This method rigorously accounts for the variability among multiple measurements of the same peptide for a specific protein and across all peptides for the same protein, the parameters σ2 and τ2, respectively, when calculating the standard error of the ratio r. Thus, the variables r(i,j,k,l) denote the ratio of the corrected and normalized peak area of MS/MS spectrum k corresponding to identified peptide j for protein i for labeled sample l = 116.1, 117.1 by the corresponding corrected and normalized peak area for 114.1. For ease of notation in the subsequent discussion, indices i and l are removed as the proposed model is used for each protein separately and for each (116.1 and 117.1) labeled sample. These ratios are modeled in a log2 scale to overcome the fact that the ratio scale is bounded from below by 0 and by the following random one-way analysis of variance model: log2(r(j,k)) = R1R(j)1u(j,k) where R is the relative abundance of protein in the labeled samples (technically the overall mean of the modeled ratios at the protein level), R(j) is the specific effect of peptide j on the ratio r(j,k), and u(j,k) is an MS/MS spectrum effect assumed to be normally distributed with mean 0 and constant variance σ2 where σ2 is a random component. It is also assumed that the peptide effects follow a normal distribution with mean 0 and variance τ2 where τ2 is a random component. Therefore, the posited model accounts for variability of the measurements both at the spectra level and at the peptide level and allows every observed relative peak area ratio to be accounted for by the overall protein ratio, by a peptide-specific value, and by an MS/MS-specific value. To expound, iTRAQ measurements from different peptides exhibit different variability, and that is the reason the peptide effect is included in the model. Because the number of peptides identified for a particular protein is not known a priori and depends on a complicated process (search algorithm, database, etc.) it is treated as a random quantity with a certain variance τ2. This variance accounts for the extra uncertainty when calculating the main effect of interest, namely one particular treatment of the cells versus another. The parameters σ2 and τ2 are estimated using a restricted maximum likelihood method (21). Accordingly p values reported are a complicated function of σ2 + τ2 due to the additional source of uncertainty. Finally proteins with p values less than or equal to 0.1 and proteins that increased or decreased equal to or greater than 10% were utilized for biological conclusions in an attempt to be most inclusive and comprehensive.
Pathway Mapping—
Network analysis was performed on proteins found to be differentially regulated in each of the experimental conditions to draw network interactions on pathways, processes, and enzymatic reactions. After conversion of IPI accession numbers to National Center for Biotechnology Information (NCBI) RefSeq accession numbers with the European Molecular Biology Laboratory (EMBL) Protein Cross-Reference File, the list was submitted to MetaCore™ (GeneGo, New Buffalo, MI) for analysis. This program has assembled enzymatic reactions and signaling protein interactions into a series of interconnected networks that have been manually curated (22).
Multiple Reaction Monitoring Mass Spectrometry—
Dried SCX fractions were reconstituted with 20 μl of 0.1% TFA in water. Reverse phase chromatography on line to MRM mass spectrometry was carried out using a NanoSpray source on a 4000 Q TRAP hybrid triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems). On-line chromatographic peptide separations were completed with a Tempo nano-LC instrument (Applied Biosystems) equipped with a 75-μm-diameter C18 PepMap reverse phase column (LC Packings, Bannockburn, IL) and eluted with gradients of 3–30% acetonitrile with 0.1% formic acid. MRM transitions were acquired at unit resolution in both the Q1 and Q3 quadrupoles to maximize sensitivity and used to trigger MS/MS in ion trap mode. Q1 MRM transitions were taken directly from global proteomics data previously obtained emphasizing those peptides that distinguish protein isoforms. MRM Q3 targets were initially predicted for peptides of interest in silico with MIDAS Workflow Designer (Applied Biosystems) for each tryptic fragment approximately conforming to the following rules. (i) y ions were preferred over b ions. (ii) A triply charged parent ion mass was assumed if the sequence contained a histidine residue. (iii) The Q3 fragment ion was to have greater mass than the selected Q1 parent ion. (iv) If the sequence contained a proline residue, the y ion created from fragmentation N-terminal to the proline was selected as a Q3 ion. All tandem mass spectra obtained during this MRM assay development were then analyzed using ProteinPilot Software (version 2.0, Applied Biosystems). Peptides that were confidently identified by the database search algorithm were further examined manually to refine and improve upon the existing Q3 target determined experimentally from the full-scan tandem mass spectrum. This process was repeated iteratively until each protein of interest had at least three unique peptides confidently identified and each peptide had three different specific MRM transitions. Once each peptide MRM was confirmed and the reverse phase retention time was noted, a quantitative series of MRMs was designed with the same Q1 target for each peptide parent ion, whereas additional Q3 MRM targets were added for the iTRAQ reporter groups ions (m/z 114.1 for control, 116.1 for noggin treatment, and 117.1 for BMP4 treatment). Quantitative MRM peaks were integrated using the peak area from the extracted ion chromatogram with tools in MultiQuant Software (version 1.0, Applied Biosystems). These peak areas were then averaged across multiple replicate analyses, and a coefficient of variation with p value was calculated.
Availability—
The data associated with this study may be downloaded from the ProteomeCommons Tranche system. The following hash code may be used to indicate exactly what files were published as part of this study's data set, and the hash code may also be used to check that the data have not changed since publication: 4UnPjSGXim01ALPA/t33bhnKAmOSoz1WrKx8rzC5nmJnOXyGQAp5uenJNP8CqwKrc2vn1winXU0MbvKenOuq6Mpuno4AAAAAAone4w=. Included Scaffold result files can be visualized with the free Scaffold viewer from Proteome Software Inc.
RESULTS
Defining the Culture System—
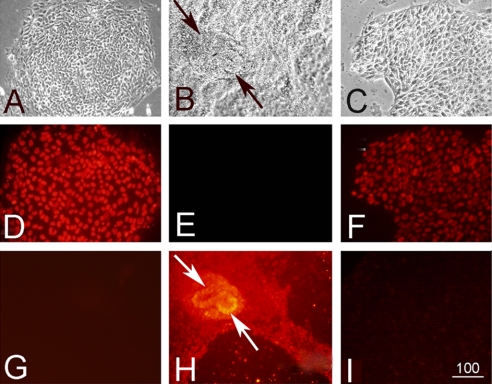
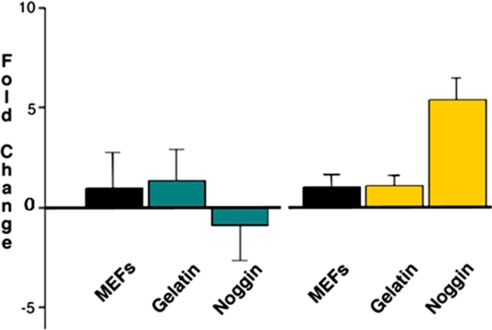
Removal of BG01 hESCs from MEFs onto gelatin-coated plates in medium supplemented with KSR and additional supplements produced little change in hESC morphology (supplemental Fig. 1A compared with Fig. 1A). Additionally these cells continued to show widespread expression of OCT3/4, a marker of pluripotent hESCs (Fig. 1D). As a control, cells were exposed to secondary antibody alone to demonstrate specificity of primary antibodies (supplemental Fig. 1B). Exposure to noggin or BMP proteins promoted differentiation as indicated by down-regulation of OCT3/4 expression (Fig. 1, E and F). After treatment with recombinant noggin protein, cells began to differentiate into primitive neurons as indicated by the expression of SOX3 (Fig. 1H; secondary antibody-Cy3 (red)), which did not occur in control cultured conditions (Fig. 1G) or after BMP4 treatment (Fig. 1I). Quantitative PCR analysis in Fig. 2 demonstrates not only the minimal effect of changing culture conditions indicated by the negligible change in expression of Oct3/4 and Sox3 but also the decreased expression of Oct3/4 and increased expression of Sox3 after 7 days of noggin treatment in these new culture conditions. These results demonstrate that exposure to noggin protein induced widespread primitive neural differentiation of these cells.
Fig. 1.
A–C, phase-contrast micrographs illustrating the appearance of BG01 hESCs growing in control conditions (A), in the presence of noggin protein (B), or in the presence of BMP4 (C). D–F, immunohistochemical localization of OCT3/4. Cells grown in control conditions continue to express high levels of OCT3/4 (D; Cy3-conjugated secondary antibody (red fluorochrome)) in contrast with very little OCT3/4 in the presence of noggin (E) and decreased levels of OCT3/4 in the presence of BMP4 (F). G–I, localization of SOX3. There was little expression of the neural progenitor marker SOX3 in control (G) or BMP4-exposed cultures (I), but there was widespread SOX3 immunoreactivity in noggin-exposed cells (H). In the noggin-treated cultures, there were neural rosettes that could be identified using phase-contrast optics (B, arrows) and immunohistochemical localization of SOX3 (H, arrows). Scale bar, 100 μm.
Fig. 2.
Quantitative RT-PCR of Oct3/4 and Sox3, markers of pluripotency and neural differentiation, respectively, before and after treatment with noggin protein. There was little effect on differentiation observed upon transfer from MEFs to gelatin in both markers; however, decreased expression of Oct4 and increased expression of Sox3 was observed after noggin exposure. Error bars represent measurement of three replicates.
After 7 days of culture in control conditions or treatment with either noggin or BMP4, three biological replicates of cells were harvested for total protein lysate. Aliquots of 100 μg of proteins from each replicate treatment condition were digested with trypsin and labeled with the iTRAQ reagents in a triple-triplex manner. Labeled and digested peptides from each replicate sample were then combined equally (w/w). This provided sufficient material for three analytical replicate analyses of each biological replicate. A total of nine experimental replicates were then separated sequentially by cation exchange and reverse phase chromatography and then analyzed by mass spectrometry.
Differentially Expressed Proteins during Lineage-specific hESC Differentiation—
A total of 45,000 spectra were generated from the nine replicate experiments and analyzed for peptide identification with three database search algorithms (Mascot, SEQUEST, X!Tandem) utilizing a decoy database for determination of FPR. Only confidently identified peptide sequences passing a filter of a 1% FPR were included in protein identifications. Table I shows the number of expected incorrect, i.e. reverse sequence, peptides identified for several different FPRs. By sorting the data by decreasing algorithm threshold, e.g. Xcorr, the calculated FPR was then correlated with the algorithm-specific threshold and number of assumed correct peptide identifications. The following thresholds were utilized for positive peptide sequence identifications that represent a 1% FPR for each database search algorithm: SEQUEST: ΔCn ≥ 0.228 and Xcorr ≥ 4.75; Mascot: identity score ≥41.8 and ion score ≥31.9; and X!Tandem: a log(e) ≥1.89. These high confidence peptides were then collapsed into a protein list using the ProteinProphet algorithm integrated into Scaffold. There were 561 unique proteins identified by all three search algorithms with a minimum of two identified peptides. An additional 15, 26, and 35 proteins were identified independently by SEQUEST, Mascot, and X!Tandem, respectively, giving a total of 603 unique proteins identified by at least two search algorithms. These calculations and this listing of protein identifications are available as a Scaffold result file in the public data repository Tranche that can be viewed using the freely available Scaffold viewer. For comparison, Table II shows the number of protein identifications for each algorithm for both a 1 and 5% peptide FPR when the minimum number of peptides per protein was decreased from 2 to 1. Then for further comparison, the number of protein identifications for two different PeptideProphet probabilities, 95 and 90%, with a minimum of one and two peptides per protein is also included.
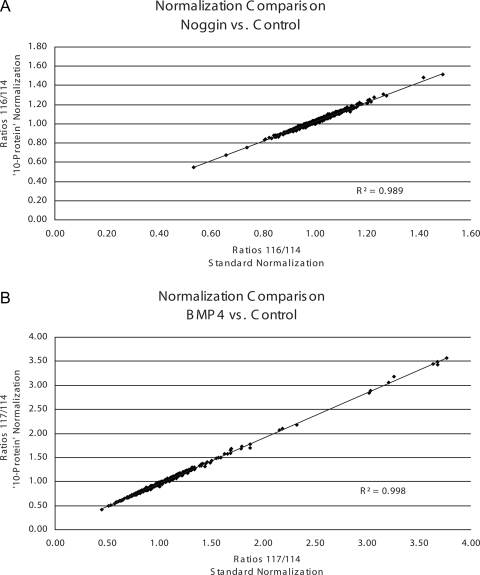
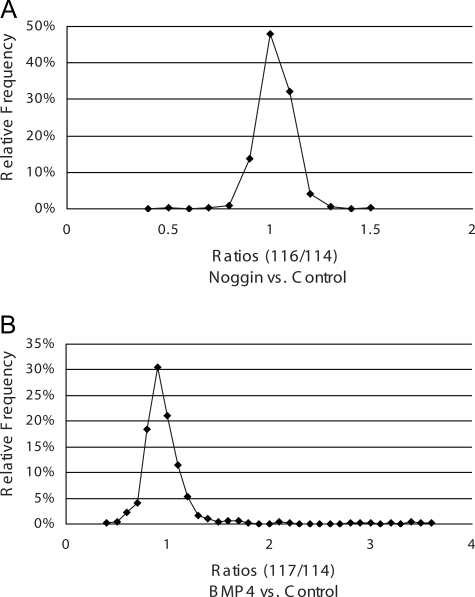
iTRAQ reporter group areas from spectra that passed the 1% peptide FPR were extracted for quantification analysis. From the total of ∼45,000 spectra, 35,874 spectra resulted in a peptide sequence identification passing the 1% FPR. Specifically 11,991, 9868, and 14,015 spectra were identified by SEQUEST, Mascot, and X!Tandem, respectively. These extracted spectra were then collapsed to remove multiple entries, i.e. where more than one search engine gave the same peptide sequence identification for the same spectra. However, multiple different spectra leading to the same peptide sequence identification were included. This resulted in 15,736 unique spectra for quantification analysis. Initially normalization for quantification was completed in a standard manner (18) where 85% of the proteins were not expected to change; however, this paradigm was not expected for this experimental system. Therefore, quantification analysis was completed and normalized against 56 peptides scanned a total of 1096 times from 10 proteins that were not expected to change. The following 10 proteins were chosen for normalization: 40 S ribosomal protein S 15, 60 S ribosomal protein L18, cytoplasmic actin, citrate synthase, cytochrome c, DNA ligase 1, glyceraldehyde-3-phosphate dehydrogenase, nuclear pore complex protein Nup133, phosphoglycerate kinase 1, and triose-phosphate isomerase 1, all of which did not change with standard normalization. Furthermore plotting and comparing the ratios of the two different normalization methodologies and calculating the correlation coefficients (shown in Fig. 3) confirmed strong consistency between the two methodologies. In both cases, however, the total number of proteins calculated as differentially regulated by the same criteria (≤/≥10%, p value ≤0.1) increased. For instance, using the standard normalization strategy, only 36 proteins were calculated to be differentially expressed in the noggin treatment group. The “10-protein” normalization strategy revealed 53 proteins as differentially expressed. This overall increase in the number of differentially expressed proteins was also seen in the BMP4 treatment group, i.e. 182 proteins with the “standard” quantification methodology compared with 206 proteins with the 10-protein strategy. Of note, the 10-protein listing is inclusive of the standard quantification listing; thus, we have only added to the number of targets that could be utilized for biological conclusions and follow-up studies. Furthermore these seemingly biologically insignificant cutoffs were used in attempts to create the most comprehensive listing for biological conclusions and follow-up studies. Therefore, utilizing the 10-protein normalization methodology, 34 proteins were calculated to be either increased or decreased ≥10% (p ≤ 0.1 in noggin versus control), whereas 187 proteins were calculated to be differentially expressed ≥10% (p ≤ 0.1 in BMP4 versus control). These listings of differentially expressed proteins are shown in supplemental Tables 1 and 2, respectively, and show a general Gaussian distribution of expression (shown in Fig. 4). An additional 19 proteins were differentially regulated by both treatments, including two that increased in both treatments, six that decreased in both treatments, and 11 that were differentially regulated in opposite directions as shown in supplemental Table 3. Of the 11 differentially regulated in opposite directions, only one protein was decreased in noggin treatment and increased in BMP4 treatment, facilitated glucose transporter membrane 1. The other 10 proteins were increased by noggin treatment and decreased by BMP4 when compared with control.
Fig. 3.
Comparison of two different quantification normalization methods: standard methodology, which assumes 85% of the identified proteins do not change, and directed protein methodology against a group of 10 proteins not expected to change biologically. Both methods were used to normalize the global proteomics quantification data. A shows the comparison of both normalization methods for the ratios of iTRAQ reporters 116 versus 114 (noggin versus control). B shows the comparison for the ratios of iTRAQ reporters 117 versus 114 (BMP4 versus control). Correlation coefficients are reported on each plot showing consistency between methodologies.
Fig. 4.
Frequency of relative expression ratios using directed protein normalization. Shown is the distribution of relative protein expression ratios for noggin treatment versus control (A) and BMP4 treatment versus control (B). Both treatment conditions show Gaussian distributions indicating very little overall change in differential expression and a shift in the general direction of decreased from control.
Validation of Targets with Orthogonal Methodologies—
Quantitative mRNA analysis was completed for the two CRMP isoforms, nuclear autogenic sperm protein (NASP), and cytokeratin-8 in both treatment groups compared with control. Results are shown in Tables III and IV for noggin and BMP4 treatment, respectively. All qRT-PCR measurements for the BMP4-treated cells compared with control (Table IV) were in concordance with regard to the direction of expression changes with the protein quantification results, although the magnitude of this direction of expression changes was found to be much more pronounced with the qRT-PCR results. However, in the noggin treatment group (Table III), mRNA expression and protein expression were not in correlation for cytokeratin-8, NASP, and CRMP2. For cytokeratin-8, both proteomics measurements indicated very little change when compared with control (0.92- and 1.02-fold) for global and targeted iTRAQ MRM analyses, respectively, whereas the qRT-PCR demonstrated a significant decrease in expression (5.28-fold). Western blotting (shown in supplemental Fig. 2) demonstrated concordance with the proteomics experiments; there was very little change in cytokeratin-8 protein expression. The NASP protein expression showed a very slight increase (1.11-fold) in the global proteomics analyses and no change (1.00-fold) in the iTRAQ MRM analyses, whereas qRT-PCR demonstrated again a significant decrease in expression (3.14-fold). Additionally CRMP2 results showed a slight increase in the global proteomics results (1.17-fold) that was not corroborated in the targeted iTRAQ MRM (1.05-fold) and qRT-PCR (1.10-fold) results as both MRM and qRT-PCR indicated no change. However, the CRMP4 results across four methodologies were in concordance (1.21- and 1.25-fold increases for global and MRM analyses, respectively) and shown to be increased in Western blotting, but a more pronounced increase (1.55-fold) was shown in the qRT-PCR findings.
Table III.
Comparison of quantification results between global proteomics experiments, quantitative RT-PCR, and targeted MRM experiments for noggin treatment versus control in hESCs
Ranges for qRT-PCR are shown in parentheses. N/A, not applicable.
| Protein | Ratio noggin vs. control
|
||
|---|---|---|---|
| Global | iTRAQ MRM | qRT-PCR | |
| Cytokeratin-8 | 0.92, p = 0.33 | 1.02, p = 0.25 | ↓5.28, S.D. = 0.8 (↓3.12–↓8.92) |
| Tubulin β-III | 1.06, p = 0.70 | 1.08, p = 0.24 | N/A |
| CRMP2 | 1.17, p = 0.09 | 1.05, p = 0.10 | ↓1.10, S.D. = 0.8 (↓1.66–↑1.38) |
| CRMP4 | 1.21, p < 0.00 | 1.25, p = 0.03 | ↑1.55, S.D. = 0.5 (↑1.08–↑2.22) |
| NASP | 1.11, p = 0.20 | 1.00, p = 0.47 | ↓3.14, S.D. = 0.6 (↓2.11–↓4.67) |
| MMIF | 1.16, p = 0.20 | 1.05, p = 0.10 | N/A |
Table IV.
Comparison of quantification results between global proteomics experiments, quantitative RT-PCR, and targeted MRM experiments for BMP4 treatment versus control in hESCs
Ranges for qRT-PCR are shown in parentheses. N/A, not applicable.
| Protein | Ratio BMP4 vs. control
|
||
|---|---|---|---|
| Global | iTRAQ MRM | qRT-PCR | |
| Cytokeratin-8 | 3.44, p < 0.00 | 6.93, p < 0.00 | ↑64.74, S.D. = 0.4 (↑48.86–↑85.80) |
| Tubulin β-III | 0.80, p < 0.00 | 0.58, p < 0.00 | N/A |
| CRMP2 | 0.72, p < 0.00 | 0.55, p = 0.03 | ↓4.19, S.D. = 0.4 (↓3.25–↓5.40) |
| CRMP4 | 0.71, p < 0.00 | 0.32, p = 0.02 | ↓4.39, S.D. = 0.3 (↓3.51–↓5.48) |
| NASP | 0.85, p = 0.02 | 0.57, p = 0.02 | ↓5.34, S.D. = 0.2 (↓4.58–↓6.22) |
| MMIF | 0.95, p = 0.60 | 0.71, p = 0.10 | N/A |
Pathway Membership of Differentially Regulated Proteins—
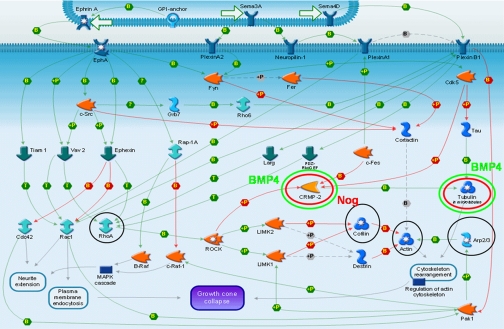
The proteins differentially regulated in both treatment groups versus control were analyzed in MetaCore, a module of GeneGo, a pathway analysis program. The protein listing was involved specifically and with high confidence in the receptor-mediated axon growth repulsion and the myelin-associated glycoprotein-dependent inhibition of neurite outgrowth pathways. Shown in Fig. 5 is the receptor-mediated axon growth repulsion pathway map. Circled in black are proteins that were identified in the global proteomics experiment but found not to be differentially regulated. Circled in green are two proteins, CRMP2 and tubulin β-III, that were found to be decreased in expression in the BMP4 treatment group; these two proteins, circled in red, were also found to be increased in the noggin treatment group.
Fig. 5.
Reconstructed pathway for receptor-mediated axon growth repulsion using GeneGo. Circled in black are proteins identified in these studies; those circled in red increased in expression, whereas those circled in green decreased in expression significantly in the treatment condition indicated. Nog, noggin; MAPK, mitogen-activated protein kinase; GPI, glycosylphosphatidylinositol; ROCK, Rho kinase; GEF, guanine nucleotide exchange factor. +P, phosphorylation; B, binding; T, transformation.
Verification of Differentiation Markers with MRM—
Proteins chosen for quantitative MRM method development fell into four general categories: (i) targets found in global experiments by only two of the three database search algorithms (macrophage migration-inhibitory factor (MMIF), hepatoma-derived growth factor (HDGF), nestin, signal transducer and activator of transcription (STAT3), and alkaline phosphatase); (ii) targets found in global experiments with high confidence but calculated to have non-significant quantification results (cytokeratin-8, tubulin β-III, NASP, myristoylated alanine-rich C-kinase substrate (MARCKS), and MARCKS-related protein (MLP)); (iii) targets found in global experiments as “one-hit wonders” with biological significance (reticulon-3, reticulon-4, isoforms 4 and 9 of mothers against decapentaplegic (SMAD4 and SMAD9), and the polycomb protein SUZ12); and (iv) targets with biological significance but that were not found in global experiments (SOX3 and OCT4).
Three peptide MRMs were designed and validated on five unique peptides for cytoskeletal keratin type II isoform 8; four peptides each for tubulin β-III, CRMP4, and NASP; three peptides each for CRMP2 and MMIF; and two peptides each for HDGF and MARCKS. Initially each in silico designed peptide MRM transition was confirmed empirically for the correct peptide sequence by obtaining the full-scan tandem mass spectrum and correct database search identification. The chromatographic peak shape of each peptide MRM transition was then assessed to ensure good resolution and therefore the ability to obtain good LC peak integration (Fig. 6). Quantification MRM transitions were then added to the method by adding additional Q3 targets for the three iTRAQ reporter ions utilizing the same peptide MRM Q1 targets permitting relative quantitation information between the three different treatment conditions.
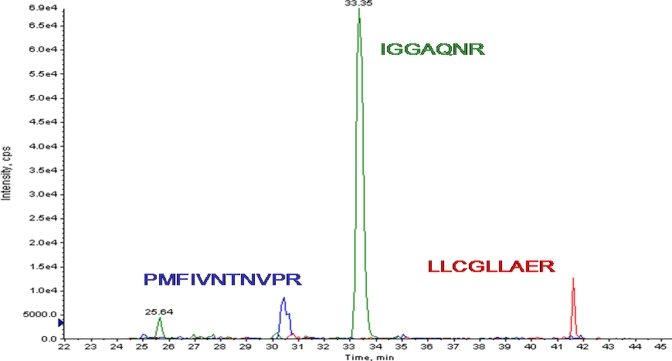
Fig. 6.
Extracted peptide MRM ion chromatograms. Each protein measured with MRM must contain at least three MRMs for each of three different peptide sequences sufficiently resolved chromatographically. The panel shows extracted ion chromatograms for three different peptides with good resolution representing the MMIF protein. cps, counts/s.
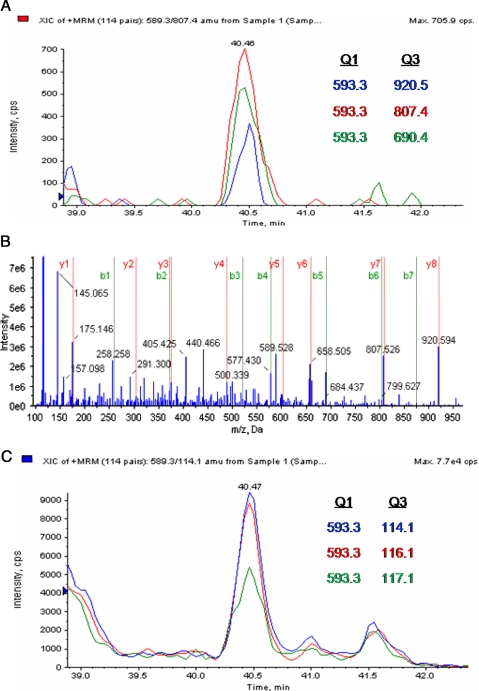
During global proteomics analysis, MMIF was identified with only two confidently sequenced peptides by only two of the three database search algorithms fulfilling criteria i for MRM design. Those two peptides were scanned by the mass spectrometer 18 times, and those 18 spectra were utilized for quantification analysis. Unfortunately the calculation of differential expression indicated a non-significant relative change, i.e. slightly increased, with a ratio of 1.16 but with a non-significant p value (p = 0.2) in the noggin treatment group and a ratio of 0.95 with a non-significant p value (p = 0.6) in the BMP4 treatment group. However, MRM analysis allowed an additional peptide sequence to be identified and more confident quantification results: 1.05-fold (p = 0.1) in noggin treatment and 0.71-fold (p = 0.10) in BMP4 treatment. Fig. 7 shows an example of the MRM work flow for identified peptide LLCGLLAER in the MMIF protein. Fig. 7A shows three extracted and validated peptide MRMs with Q1 and Q3 targets noted. This peptide was identified as the doubly charged ion 598.3 m/z. Q3 targets were designed and validated for y8 ion 920.5 m/z, y7 ion 807.4 m/z, and b5 ion 690.4 m/z. Fig. 7B shows the full-scan tandem mass spectrum indicating the presence of a complete y ion series and an almost complete b ion series, thereby confidently identifying the sequence of this peptide. Three additional MRM transitions were then added to the method with the doubly charged peptide parent ion 598.3 m/z as Q1 target and the iTRAQ reporter ions as Q3 targets. Fig. 7C shows the additional extracted chromatogram for the quantitative MRMs for this peptide on which areas were calculated for relative quantification. During this MRM analysis, three unique peptides for MMIF were identified and quantified a total of six times. This quantitative MRM analysis revealed with more significance the relative change with a ratio of 1.05 (p = 0.1; coefficient of variance, ±19%) and 0.71 (p = 0.1; coefficient of variance, ±17%) in the noggin and BMP4 treatment groups compared with control, respectively. In summary, with global proteomics experiments only two of the three search algorithms identified MMIF, and the relative differential expression did not reveal a confident assessment; however, MRM experiments demonstrated with more confidence the identification of the protein and the calculation of relative expression difference.
Fig. 7.
Peptide LLCGLLAER sequence from MMIF. A shows an extracted ion chromatogram (XIC) for three different sequence ion MRMs for peptide ion 589.3 m/z. B shows a full-scan tandem mass spectrum confirming the sequence. C shows the MRM transitions used for relative quantification. Extracted ion chromatograms are shown for the same peptide parent ion with same LC retention time, but iTRAQ reporter ions were used as targets for Q3. cps, counts/s.
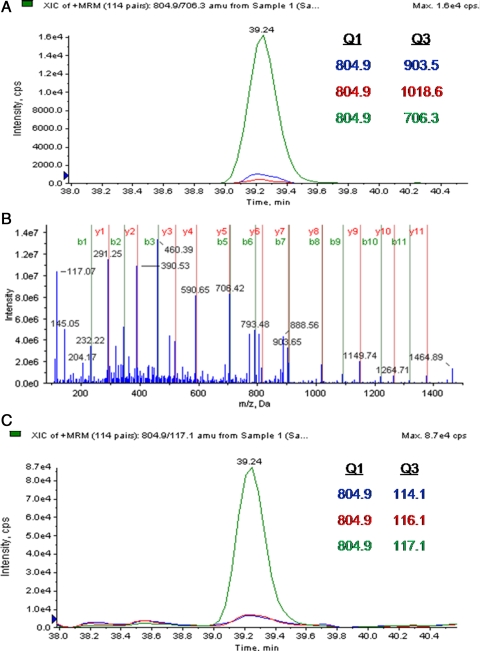
Fig. 8 illustrates another example of the MRM work flow for the identified peptide SLDMDSIIAEVK in cytokeratin-8. In this example, the cytokeratin-8 protein was identified in global proteomics experiments with 23 peptides, some of which are redundant for other cytokeratin isoforms. These peptides were scanned 107 times and were quantified as being unchanged in the noggin treatment group (0.92, p = 0.33) and significantly increased in the BMP4 treatment group (3.45, p ≤ 0.00). In contrast, during MRM analysis we identified five unique, isoform-specific peptides and quantified these peptides eight times that when calculated illustrates concordant and more statistically confident results, i.e. unchanged in the noggin treatment group (1.02-fold, p = 0.25) and considerably overexpressed (6.93-fold, p < 0.00) in the BMP4 treatment group.
Fig. 8.
Peptide SLDMDSIIAEVK sequence from cytokeratin-8. A shows an extracted ion chromatogram (XIC) for three different MRMs for peptide ion 804.9 m/z. B shows a full-scan tandem mass spectrum confirming the sequence. C, quantitative MRMs showing extracted ion chromatogram for the same peptide parent ion with same LC retention time, but iTRAQ reporter ions were used as targets for Q3. cps, counts/s.
MRM analyses for an additional six proteins were completed in this manner, emphasizing additional peptide identification information as well as isoform-specific quantification measurements. Tables III and IV outline for comparison the quantification results for the global proteomics screening experiments, the directed MRM studies, and the qRT-PCR results. In all cases for BMP4 treatment, the MRM results showed concordant direction of expression albeit a greater magnitude of that differential expression. Two proteins in the noggin treatment group showed similar results regarding almost no protein expression change between the global and MRM technologies, cytokeratin-8 and tubulin β-III. Three additional proteins in the noggin-treated cells (CRMP2, NASP, and MMIF) were shown to have a greater expression change in the global proteomics experiments compared with the targeted MRM analyses that showed no significant change, whereas one protein (CRMP4) showed concordant results for both proteomics technologies.
DISCUSSION
To address protein dynamic range issues and potential artifacts of co-culture, a model system was developed to decrease sample complexity and avoid potential confounding cell types/proteins. Typically hESCs grow on a feeder layer of irradiated MEFs. This practice introduces three particular challenges for proteomics studies. First, the potential to carry over mouse proteins introduces complexity with sequence homology from the different species. Second, MEFs are much greater in size and weight than hESCs, adding to the already challenging large dynamic range of protein concentration: MEF proteins will be more concentrated than hESC proteins, thereby masking the measurement and identification of proteins from hESCs. The final concern is that added protein (either noggin or BMP4 in these experiments) may cause MEFs to express proteins that affect hESCs in an unpredictable and uncontrolled manner despite the fact that MEFs were irradiated. To eliminate these problems, we used a feeder-free culture system with defined medium.
Removal of the hESCs from irradiated MEFs to a defined culture medium on a substrate of gelatin did not change the morphological appearance shown in supplemental Fig. 1A compared with Fig. 1A. Furthermore the expression of Oct3/4 and Sox3 also did not change upon transfer as shown with qRT-PCR data in Fig. 2. As is well documented for mESCs, noggin promoted neural differentiation of hESCs. Fig. 1 illustrates that when these cells were treated with recombinant noggin protein (Fig. 1, B, E, and H) they adopted a neuronal morphology (Fig. 1B), decreased expression of OCT3/4 (Fig. 1E), and expressed the primitive neuronal marker SOX3 (Fig. 1H). Furthermore treatment of these cells with BMP4, as indicated by lack of SOX3 staining (Fig. 1I), did not induce neuronal differentiation. These results confirmed and demonstrated that in hESCs, as in mESCs, exposure to noggin protein induces widespread neural differentiation that does not occur with BMP4 exposure (2, 3, 5, 23, 24).
After completing the proof-of-principle experiments in the new culture conditions, a comprehensive global quantification proteomics experiment was undertaken. To compensate for biological variation, three biological replicates were conducted. Additionally three analytical replicates for each of the three biological replicates were completed, creating an ample, nine-replicate data set for analysis. Approximately 45,000 tandem mass spectra were analyzed thoroughly with three database search algorithms, SEQUEST, Mascot, and X!Tandem. Although these three algorithms are not orthogonal, they do contain subtle differences in their heuristic approaches. Each also uses different threshold approaches for determining correctness of peptide identification. Correlating one threshold in one algorithm to a threshold for a different algorithm is an enormous challenge. Thus, a target decoy database strategy was utilized (20) to allow comparison of the different thresholds. In this strategy, the reversed sequences of a protein database are concatenated to the forward sequences. This permits the calculation of an FPR using the following equation: FPR = 2 × (FP/(TP + FP)) where FP is the false positive peptide identification (reverse sequence-identified) and TP is the true positive peptide identification assumed to be correct by identification of a forward sequence. The integer 2 is used to compensate for the doubling in size of the database. Each peptide identified as a reverse sequence is assumed incorrect and of random chance irrespective of database search algorithm. In this way, we can calculate the overall FPR for each algorithm specific to this data set and compare each database search algorithm results with one another. Table I shows respective estimations of the number of correct peptide identifications at a calculated FPR for each algorithm. Then each confidently identified peptide and associated protein identification were analyzed for relative quantification utilizing in-house statistical comparison. Of the ∼45,000 obtained spectra, 15,736 resulted in unique peptide identifications for quantification analysis. These peptide sequences collapsed into 603 unique proteins with a minimum of two peptides per protein with quantification data. Of these identified and quantifiable proteins, 53 were shown to be differentially express during noggin treatment, and 206 were shown to be differentially expressed during BMP4 treatment.
Unfortunately the combination of methodologies and iTRAQ chemical labeling coupled with the 4800 MALDI-TOF/TOF instrumentation can result in compression of the iTRAQ reporter group ratios. This is suspected to be caused by a large timed ion selector window allowing low level peptide ions of similar masses to enter the collision cell and be co-fragmented with the target ion. Consequently iTRAQ reporter groups originating from different peptides yet close in mass are measured simultaneously. Those nonspecific data then contribute to the ratios calculated, resulting in an overall averaging or compression effect. This effect is particularly acute for high complexity samples and low resolution chromatographic separations. Due to this phenomenon, the cutoffs for the magnitude of differential expression as well as the significance of calculation were lessened in significance to a change of ≥/≤10% and a p value of ≤0.1. This allowed us to consider the most comprehensive listing of protein quantifications for biological conclusions that could be verified and validated by alternative technologies and methodologies.
Of the proteins we confidently identified and quantified with global proteomics methodologies, many had been reported previously (4, 25–27), providing a degree of assurance regarding the approach. For example, following noggin treatment, the neural cell-specific proteins tubulin β-III and the CRMP isoforms were identified and in the case of CRMP4 found to be increased as expected (4, 28, 29). Following BMP4 treatment, these specific neural proteins were quantified as being decreased as expected. This validates not only our new culture conditions and the species similarities with regard to noggin and BMP4 exposure in hESCs and mESCs but also the quantitative proteomics methodologies. CRMP2 and CRMP4 (also known as dihydropyrimidinase-related proteins DPYSL2 and DPYSL3, respectively) were identified in noggin treatment and found to be significantly decreased with BMP4 treatment. CRMP2 is reported to be phosphorylated by Rho (identified in this study) that in turn regulates the phosphorylation of myosin light chain (also identified in this study) resulting in actinomyosin contractility (28). During the development of the nervous system, nerve growth cones play a central role in axon guidance (30). Overexpression of CRMP2 has been shown to promote axonogenesis, whereas expression of a dominant-negative form or knockdown of CRMP2 suppresses axon formation (31). Although much is known about CRMP2, little is known about CRMP4; however, its localization is mainly in neurogenic regions of the central nervous system (32, 33), and it is strongly expressed in early embryonic postmitotic neural cells (34). It was recently shown that CRMP4 is cleaved by calpain during excitotoxicity and oxidative stress; this may have a significant impact on its interaction with actin and its assembly and in turn on growth cone dynamics (35). Other interesting neural proteins confidently identified in this global proteomics screen include MLP and nestin. In mice, mutated MLP results in severe neural tube defects, including exencephaly, spina bifida, and a tail flexion anomaly (36). Nestin, long known to be highly expressed in neural stem cells, is a marker of early neural differentiation (37–39). Both of these proteins were confidently identified in both treatment conditions but not differentially regulated in the noggin-treated cells as expected. However, both proteins were found to be significantly decreased in the BMP4 treatment group compared with control, further confirming as expected that BMP4 treatment causes an inhibition of neural differentiation.
Proteins differentially expressed in the BMP4 treatment group included HDGF, which was decreased compared with control, substantiating reports that this protein is present in undifferentiated hESCs (13, 40). Additionally we identified differential regulation of the germ cell factor NASP. The function of NASP is to transport and exchange H1 histones into nuclei with DNA (41). Null mutation NASP−/− is lethal at the blastocyst stage when hESCs are obtained (41). This protein was found to be decreased in expression during BMP4 treatment, supporting an effect of BMP4 treatment in lineage differentiation and countering the suggestion of its positive effect on self-renewal. Cytokeratin-8 and -18 were also identified in this work to be increased after BMP4 treatment and slightly decreased after noggin treatment compared with the undifferentiated control. The subset of cytokeratins that an epithelial cell expresses depends mainly on the type of epithelium, the moment in the course of terminal differentiation, and the stage of development. Cytokeratin-8 and -18 are nonspecific markers of a wide range of epithelial tissues originating from ectodermal differentiation but are not expressed by neural cells, indicating that BMP4 treatment indeed caused differentiation away from neural cell types and toward an epidermal endoderm lineage.
Overall there were a relatively low number of total proteins differentially regulated in both treatment groups as shown in supplemental Tables 1 and 2. Furthermore their predominant direction was decreased from control. Along with the Gaussian distribution of protein expression shown in Fig. 4, these results support the suggestion that hESCs normally express most of the proteins required to specify each lineage, i.e. a primed state. A common feature among stem cells is the low expression level of a large number of genes involved in multilineage differentiation (42, 43) that may be required for rapid differentiation (44). Therefore, it is not surprising that treatment with BMP4 or noggin promotes commitment of the hESCs to a particular lineage and reduces the expression of those proteins no longer needed for differentiation to other lineages.
Substantiating this genetic evidence reported previously, our study showed that BMP4 exposure decreases the relative expression of MLP when compared with control hESCs, whereas noggin treatment results in little change from the control pluripotent cells. MLP is highly expressed in the developed brain and not found in other tissues (36). Therefore, it is not surprising that we found its expression to decrease upon BMP4 treatment. Furthermore the fact that there was no relative change in expression after noggin treatment compared with control implies a requirement for MLP in developing neural tissues at the earliest stages of development. Increased MLP expression may not be required for undifferentiated hESCs and or early neural progenitors but is similarly expressed in both cell types prior to commitment to terminal neural differentiation. These data and others examples not provided support the hypothesis that pluripotent stem cells contain not only the mRNAs but may also contain many of the proteins needed for multilineage development. However, once a cell commits toward a specific lineage, those proteins no longer needed are decreased in expression, whereas those proteins needed for that lineage remain unchanged or increase in expression.
These global proteomics data also provide insight into the comparative molecular and cellular control of differentiation of both mESCs and hESCs. To date, most studies using ESCs have been completed on mouse, Xenopus, or Drosophila. The data reported here for hESCs further substantiate that noggin treatment indeed induces differentiation toward a neural cell fate similarly to its effect on mESCs. Additionally BMP4 treatment of hESCs induced differentiation away from neural differentiation and toward an epidermal ectoderm lineage similar to reports in mESCs (23, 45) and did not promote/maintain self-renewal. This is supported both by the decreased expression of several neural cell-specific proteins (tubulin β-III, CRMP4, nestin, and MLP) and the increased expression of two key ectoderm proteins (cytokeratin-8 and cytokeratin-18). Finally the identification of several expected differentiation markers suggests that global proteomics, as a methodology, is an effective way to elucidate, in a systems biology, non-a priori manner, proteins involved in targeted differentiation.
To confirm the differentially expressed proteins, qRT-PCR was completed on four biological targets that could be utilized as biomarkers of differentiation: cytokeratin-8, a marker of epidermal ectoderm differentiation; the two isoforms of CRMP for neuronal differentiation; and the putative pluripotency marker NASP. For all molecular targets in the BMP4 treatment group, comparison between differential expression of mRNA and protein expression measured in the global proteomics screening was in complete concordance with regard to the direction of differential expression. However, there were discrepancies when comparing the mRNA and protein expression in the noggin treatment group. For instance, there was little to no change in cytokeratin-8 and NASP expression reflected in the noggin treatment group compared with control; however, a significant decrease was present in the mRNA expression. Cytokeratin-8 was found to change little in the global proteomics screening (0.92-fold, p = 0.33), and this measurement was calculated with a non-significant p value. Conversely the qRT-PCR results showed a significant decrease in mRNA expression (5.28-fold). The second discrepancy between mRNA and protein differential expression in the noggin treatment group was in the NASP protein, a protein found in postmeiotic oocytes and spermatids (41). Like cytokeratin-8, in the noggin treatment group, NASP changed little in the global proteomics screening (1.11-fold, p = 0.2). Conversely the mRNA was found to be decreased significantly (3.14-fold). Given the different kinetics between mRNA and protein frequently observed (46–48), we hypothesize that, if the treatment conditions were carried out longer, the protein expression changes would mimic mRNA expression. Or alternatively, these data simply reflect a dose-response effect. That is, although it appears that BMP4 treatment was sufficient to fully realize lineage segregation, either noggin exposure may have been high enough or exposure was sufficiently short to only cause very primitive neural differentiation. An alternative explanation is that noggin treatment promotes differentiation of a pool of primitive neurons that remain growth factor-responsive.
After analysis of these quantitative global proteomics findings and qRT-PCR, we proposed to design a high throughput methodology to measure specific biomarkers of stem cell differentiation and self-renewal. There were two specific goals in extending this study with MRM: the first was to verify the global proteomics findings (both the identity and quantity of the proteins), and the second was to develop an approach for distinguishing between different homologous protein isoforms. Prototypic examples of the MRM work flow are illustrated in Figs. 7 and 8. Upon in silico prediction of Q1 and Q3 fragments, the identification of the peptide was confirmed both by the database search algorithm Paragon (49) in the ProteinPilot Software and manual inspection. At this time, optimization of Q3 targets was completed, if needed, based on the full-scan tandem mass spectrum of the peptide sequence. The assay development phase of peptide MRM generation had the additional challenge that the desired peptides could be present in any of the salt fractions and therefore required several acquisitions across multiple fractions until the correct fraction was analyzed and data were obtained. As a consequence, most of the sample allocated was consumed in this process, preventing the in-depth study of additional potentially interesting and important protein targets. Therefore, although a substantial attempt was made to complete discovery MRM analysis on specific salt fractions and load appropriate concentrations (giving consideration to global experimentation data), many initial targets representing one-hit wonder proteins or proteins never detected in global proteomics experiments were also not detected during the MRM discovery phase. Nevertheless successful MRM transitions from multiple proteins (eight proteins, 25 peptides, and ∼75 MRMs) were obtained. With an optimized peptide MRM transitions and reproducible reverse phase retention time, quantitative MRM transitions were included in the methodology utilizing the iTRAQ reporter groups as Q3 targets.
The peptide MRM, full-scan tandem mass spectrum, and quantitative MRM for one peptide in MMIF is illustrated in Fig. 7. MMIF was chosen as a target for MRM method development because relative quantification of both treatment groups compared with control showed little differential protein expression and further was calculated to have high, i.e. non-significant, p values (Tables III and IV). This protein additionally was only identified by two of three of the database search algorithms. The MRM analysis confirmed the identity of MMIF with additional peptide sequence information and also confirmed its negligible differential expression. Another protein, HDGF, which has been associated with differentiation, was identified in the global proteomics experiments with high confidence in only two of three database search algorithms and further with only one peptide; however, MRM analysis allowed confident identification with a total of three peptides including the one previously identified. All quantitative MRM analyses on the six proteins targeted generally corroborated, at least in direction of expression, the results found during global proteomics experiments as indicated in Tables III and IV. Therefore, the use of MRM analysis for verification of peptide identifications can provide additional quantitative information and confidence in the differential expression of proteins.
The variability in the magnitude of differential expression between the global proteomics data and the MRM data shown in Tables III and IV led to the isoform interference hypothesis. This simple phenomenon arises from peptides that share sequence homology within multiple protein isoforms and can dilute the measurement of relative differential expression of any one particular protein isoform. This observation is exemplified in the CRMP isoforms. Although CRMP2 is a splice form of CRMP4, they share 75.8% homology of the overlapping sequences. Global proteomics experiments identified peptides that were both unique and in common to each isoform. It is our contention that those peptides identified in common to both isoforms should not be used in the computation of the overall protein ratio as it would result in inaccurate results. That is, the redundant peptides shared between protein isoforms contribute a concentration-weighted ratio of all expressed isoforms to the overall specific isoform. Therefore, MRM transitions were specifically designed and targeted to include only those unique sequence peptides that distinguish isoforms and provide isoform-specific quantification results. Because MRM transitions were designed for non-redundant peptides, the magnitude of differential expression was more pronounced in several cases. Additionally we utilized MRM to distinguish between the tropomyosin 2 and 4 isoforms and identified additional peptides to confirm the identification of MARCKS (data not shown). Taken together, these results confirm that an alternative methodology, although not orthogonal, can be used to confirm and enhance global proteomics results. However, because of the gross lack of concordance with the noggin treatment group with regard to the proteomics results and the qRT-PCR for several targets, Western blotting was carried out for both cytokeratin-8 and CRMP4 because isoform-specific antibodies were commercially available. These results, shown in supplemental Fig. 2, show that the protein expression of cytokeratin-8 showed little change in expression, which is completely contrary to the qRT-PCR results, although there was a slight increase in CRMP4, which is in concordance with both global and qRT-PCR results. Both of these Western blotting results authenticate both the global and the targeted iTRAQ MRM proteomics analyses. Unfortunately further isoform-specific antibodies commercially available did not produce the specificity needed to be conclusive (data not shown).
CONCLUSIONS
We have developed a cell culture system in which hESCs can be investigated to identify the specific mechanisms implicated during self-renewal and targeted differentiation using a systems biology approach without confounding experimental conditions. This approach allowed the identification of several lineage-specific proteins and advocates species similarity between mESCs and hESCs with regard to the role of BMP4 signaling in differentiation. These data also support the priming hypothesis of embryonic stem cells; that is, ESCs express, albeit at low levels, the mRNA and proteins needed to differentiate into every lineage. However, upon lineage segregation, those messages and proteins no longer needed for alternative lineages decrease in expression or otherwise become eliminated as shown by the predominant decreased direction of differential protein expression in these experiments and specific examples such as the CRMP isoforms, tubulin β-III, and MLP.
At its core, however, this work represents the successful transition from discovery proteomics to target verification with multiple reaction monitoring. Although not entirely orthogonal, the combination of proteomics platforms permitted the elucidation, identity confirmation, and quantitative verification of eight proteins that may be further utilized as biomarkers in stem cell biology. Two of these markers, tubulin β-III and cytokeratin-8, were previously recognized as markers for neuronal and epithelial differentiation, respectively, and demonstrate the validity of our culture methods and experimental design. Further we propose that NASP may be a useful marker of pluripotency, and CRMP2 and CRMP4 may be useful as markers of early neuronal differentiation. Utilizing MRM, we specifically targeted unique non-redundant peptides to increase confidence in protein identifications and for quantification of particular homologous protein isoforms. This permitted what we believe to be more accurate measurements of their differential expression. These data demonstrate the power of MRM to provide a highly selective and potentially highly sensitive, high throughput methodology, which will now be further optimized for greater sensitivity and has the capacity to facilitate the productive utilization of hESCs for regenerative medicine.
Supplementary Material
Acknowledgments
We thank Crystal Pacut and Marianna Borrman for help with hESC culture; Lisa DeBoer, Nicole Slawny, and Derrick Yang for help with qRT-PCR analysis; and Yong Li for help with Western blotting.
Footnotes
Published, MCP Papers in Press, February 26, 2008, DOI 10.1074/mcp.M700399-MCP200
The abbreviations used are: hESC, human embryonic stem cell; BMP, bone morphogenic protein; CRMP, collapsin response mediator protein; FPR, false positive rate; FP, false positive; TP, true positive; HDGF, hepatoma-derived growth factor; KSR, knock-out serum replacer; MARCKS, myristoylated alanine-rich C-kinase substrate; MEF, mouse embryonic fibroblast; mESC, mouse embryonic stem cell; MLP, MARCKS-related protein; MMIF, macrophage migration-inhibitory factor; NASP, nuclear autoantigenic sperm protein; 2D, two-dimensional; qRT-PCR, quantitative RT-PCR; α-, anti-; iTRAQ, isobaric tags for relative and absolute quantification; SCX, strong cation exchange; IPI, International Protein Index.
This work was supported by National Institutes of Health Grants 1P41 RR 018627, GM-069985, NS-04187, P20 GM-069985, NS-39438, MTTC GR-687, and MEDC GR-687. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Yanagita, M. (2005) BMP antagonists: their roles in development and involvement in pathophysiology. Cytokine Growth Factor Rev. 16, 309–317 [DOI] [PubMed] [Google Scholar]
- 2.Balemans, W., and Van Hul, W. (2002) Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev. Biol. 250, 231–250 [PubMed] [Google Scholar]
- 3.Itsykson, P., Ilouz, N., Turetsky, T., Goldstein, R. S., Pera, M. F., Fishbein, I., Segal, M., and Reubinoff, B. E. (2005) Derivation of neural precursors from human embryonic stem cells in the presence of noggin. Mol. Cell. Neurosci. 30, 24–36 [DOI] [PubMed] [Google Scholar]
- 4.Tropepe, V., Hitoshi, S., Sirard, C., Mak, T. W., Rossant, J., and van der Kooy, D. (2001) Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron 30, 65–78 [DOI] [PubMed] [Google Scholar]
- 5.Gratsch, T. E., and O'Shea, K. S. (2002) Noggin and chordin have distinct activities in promoting lineage commitment of mouse embryonic stem (ES) cells. Dev. Biol. 245, 83–94 [DOI] [PubMed] [Google Scholar]
- 6.Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., and Jones, J. M. (1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 7.Daheron, L., Opitz, S. L., Zaehres, H., Lensch, W. M., Andrews, P. W., Itskovitz-Eldor, J., and Daley, G. Q. (2004) LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells 22, 770–778 [DOI] [PubMed] [Google Scholar]
- 8.Rao, M. (2004) Conserved and divergent paths that regulate self-renewal in mouse and human embryonic stem cells. Dev. Biol. 275, 269–286 [DOI] [PubMed] [Google Scholar]
- 9.Boiani, M., and Scholer, H. R. (2005) Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 6, 872–884 [DOI] [PubMed] [Google Scholar]
- 10.Sonntag, K. C., Pruszak, J., Yoshizaki, T., van Arensbergen, J., Sanchez-Pernaute, R., and Isacson, O. (2007) Enhanced yield of neuroepithelial precursors and midbrain-like dopaminergic neurons from human embryonic stem cells using the bone morphogenic protein antagonist noggin. Stem Cells 25, 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Shea, K. S. (2004) Self-renewal vs. differentiation of mouse embryonic stem cells. Biol. Reprod. 71, 1755–1765 [DOI] [PubMed] [Google Scholar]
- 12.Yao, S., Chen, S., Clark, J., Hao, E., Beattie, G. M., Hayek, A., and Ding, S. (2006) Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc. Natl. Acad. Sci. U. S. A. 103, 6907–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Hoof, D., Mummery, C. L., Heck, A. J., and Krijgsveld, J. (2006) Embryonic stem cell proteomics. Expert Rev. Proteomics 3, 427–437 [DOI] [PubMed] [Google Scholar]
- 14.Van Hoof, D., Passier, R., Ward-Van Oostwaard, D., Pinkse, M. W., Heck, A. J., Mummery, C. L., and Krijgsveld, J. (2006) A quest for human and mouse embryonic stem cell-specific proteins. Mol. Cell. Proteomics 5, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 15.Baharvand, H., Hajheidari, M., Ashtiani, S. K., and Salekdeh, G. H. (2006) Proteomic signature of human embryonic stem cells. Proteomics 6, 3544–3549 [DOI] [PubMed] [Google Scholar]
- 16.Anderson, L., and Hunter, C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 17.Livak, K. J., and Schmittgen, T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 18.Jagtap, P., Michailidis, G., Zielke, R., Walker, A. K., Patel, N., Strahler, J. R., Driks, A., Andrews, P. C., and Maddock, J. R. (2006) Early events of Bacillus anthracis germination identified by time-course quantitative proteomics. Proteomics 6, 5199–5211 [DOI] [PubMed] [Google Scholar]
- 19.Falkner, J. A., Falkner, J. W., and Andrews, P. C. (2007) ProteomeCommons.org IO Framework: reading and writing multiple proteomics data formats. Bioinformatics 23, 262–263 [DOI] [PubMed] [Google Scholar]
- 20.Elias, J. E., and Gygi, S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 21.McCulloch, C., and Searle, S. (2001) Generalized, Linear, and Mixed Models, John Wiley & Sons, Inc., New York
- 22.Ekins, S., Nikolsky, Y., Bugrim, A., Kirillov, E., and Nikolskaya, T. (2007) Pathway mapping tools for analysis of high content data. Methods Mol. Biol. 356, 319–350 [DOI] [PubMed] [Google Scholar]
- 23.Hogan, B. L. (1996) Bmps: multifunctional regulators of mammalian embryonic development. Harvey Lect. 92, 83–98 [PubMed] [Google Scholar]
- 24.McMahon, J. A., Takada, S., Zimmerman, L. B., Fan, C. M., Harland, R. M., and McMahon, A. P. (1998) Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 12, 1438–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo, X., Ying, W., Wan, J., Hu, Z., Qian, X., Zhang, H., and He, F. (2001) Proteomic characterization of early-stage differentiation of mouse embryonic stem cells into neural cells induced by all-trans retinoic acid in vitro. Electrophoresis 22, 3067–3075 [DOI] [PubMed] [Google Scholar]
- 26.Hoffrogge, R., Mikkat, S., Scharf, C., Beyer, S., Christoph, H., Pahnke, J., Mix, E., Berth, M., Uhrmacher, A., Zubrzycki, I. Z., Miljan, E., Volker, U., and Rolfs, A. (2006) 2-DE proteome analysis of a proliferating and differentiating human neuronal stem cell line (ReNcell VM). Proteomics 6, 1833–1847 [DOI] [PubMed] [Google Scholar]
- 27.Sarkar, S. A., and Sharma, R. P. (2002) Expression of selected apoptosis related genes, MIF, IGIF and TNFα, during retinoic acid-induced neural differentiation in murine embryonic stem cells. Cell Struct. Funct. 27, 99–107 [DOI] [PubMed] [Google Scholar]
- 28.Arimura, N., Inagaki, N., Chihara, K., Menager, C., Nakamura, N., Amano, M., Iwamatsu, A., Goshima, Y., and Kaibuchi, K. (2000) Phosphorylation of collapsin response mediator protein-2 by Rho-kinase. Evidence for two separate signaling pathways for growth cone collapse. J. Biol. Chem. 275, 23973–23980 [DOI] [PubMed] [Google Scholar]
- 29.Arimura, N., Menager, C., Kawano, Y., Yoshimura, T., Kawabata, S., Hattori, A., Fukata, Y., Amano, M., Goshima, Y., Inagaki, M., Morone, N., Usukura, J., and Kaibuchi, K. (2005) Phosphorylation by Rho kinase regulates CRMP-2 activity in growth cones. Mol. Cell. Biol. 25, 9973–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aitken, A., Baxter, H., Dubois, T., Clokie, S., Mackie, S., Mitchell, K., Peden, A., and Zemlickova, E. (2002) Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem. Soc. Trans. 30, 351–360 [DOI] [PubMed] [Google Scholar]
- 31.Kawano, Y., Yoshimura, T., Tsuboi, D., Kawabata, S., Kaneko-Kawano, T., Shirataki, H., Takenawa, T., and Kaibuchi, K. (2005) CRMP-2 is involved in kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol. Cell. Biol. 25, 9920–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinn, C. C., Chen, E., Kinjo, T. G., Kelly, G., Bell, A. W., Elliott, R. C., McPherson, P. S., and Hockfield, S. (2003) TUC-4b, a novel TUC family variant, regulates neurite outgrowth and associates with vesicles in the growth cone. J. Neurosci. 23, 2815–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinn, C. C., Gray, G. E., and Hockfield, S. (1999) A family of proteins implicated in axon guidance and outgrowth. J. Neurobiol. 41, 158–164 [PubMed] [Google Scholar]
- 34.Charrier, E., Reibel, S., Rogemond, V., Aguera, M., Thomasset, N., and Honnorat, J. (2003) Collapsin response mediator proteins (CRMPs): involvement in nervous system development and adult neurodegenerative disorders. Mol. Neurobiol. 28, 51–64 [DOI] [PubMed] [Google Scholar]
- 35.Kowara, R., Chen, Q., Milliken, M., and Chakravarthy, B. (2005) Calpain-mediated truncation of dihydropyrimidinase-like 3 protein (DPYSL3) in response to NMDA and H2O2 toxicity. J. Neurochem. 95, 466–474 [DOI] [PubMed] [Google Scholar]
- 36.Stumpo, D. J., Eddy, R. L., Jr., Haley, L. L., Sait, S., Shows, T. B., Lai, W. S., Young, W. S., III, Speer, M. C., Dehejia, A., Polymeropoulos, M., and Blackshear, P. J. (1998) Promoter sequence, expression, and fine chromosomal mapping of the human gene (MLP) encoding the MARCKS-like protein: identification of neighboring and linked polymorphic loci for MLP and MACS and use in the evaluation of human neural tube defects. Genomics 49, 253–264 [DOI] [PubMed] [Google Scholar]
- 37.Bazan, E., Alonso, F. J., Redondo, C., Lopez-Toledano, M. A., Alfaro, J. M., Reimers, D., Herranz, A. S., Paino, C. L., Serrano, A. B., Cobacho, N., Caso, E., and Lobo, M. V. (2004) In vitro and in vivo characterization of neural stem cells. Histol. Histopathol. 19, 1261–1275 [DOI] [PubMed] [Google Scholar]
- 38.Hockfield, S., and McKay, R. D. (1985) Identification of major cell classes in the developing mammalian nervous system. J. Neurosci. 5, 3310–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahler, J., and Driever, W. (2007) Expression of the zebrafish intermediate neurofilament Nestin in the developing nervous system and in neural proliferation zones at postembryonic stages. BMC Dev. Biol. 7, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rathjen, J., Lake, J. A., Bettess, M. D., Washington, J. M., Chapman, G., and Rathjen, P. D. (1999) Formation of a primitive ectoderm like cell population, EPL cells, from ES cells in response to biologically derived factors. J. Cell Sci. 112, 601–612 [DOI] [PubMed] [Google Scholar]
- 41.Richardson, R. T., Alekseev, O. M., Grossman, G., Widgren, E. E., Thresher, R., Wagner, E. J., Sullivan, K. D., Marzluff, W. F., and O'Rand, M. G. (2006) Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J. Biol. Chem. 281, 21526–21534 [DOI] [PubMed] [Google Scholar]
- 42.Carter, M. G., Sharov, A. A., VanBuren, V., Dudekula, D. B., Carmack, C. E., Nelson, C., and Ko, M. S. (2005) Transcript copy number estimation using a mouse whole-genome oligonucleotide microarray. Genome Biol. 6, R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zipori, D. (2004) The nature of stem cells: state rather than entity. Nat. Rev. Genet. 5, 873–878 [DOI] [PubMed] [Google Scholar]
- 44.Niwa, H. (2007) How is pluripotency determined and maintained? Development 134, 635–646 [DOI] [PubMed] [Google Scholar]
- 45.Hogan, B. L. (1996) Bone morphogenetic proteins in development. Curr. Opin. Genet. Dev. 6, 432–438 [DOI] [PubMed] [Google Scholar]
- 46.Chen, Y. R., Juan, H. F., Huang, H. C., Huang, H. H., Lee, Y. J., Liao, M. Y., Tseng, C. W., Lin, L. L., Chen, J. Y., Wang, M. J., Chen, J. H., and Chen, Y. J. (2006) Quantitative proteomic and genomic profiling reveals metastasis-related protein expression patterns in gastric cancer cells. J. Proteome Res. 5, 2727–2742 [DOI] [PubMed] [Google Scholar]
- 47.McNicoll, F., Drummelsmith, J., Muller, M., Madore, E., Boilard, N., Ouellette, M., and Papadopoulou, B. (2006) A combined proteomic and transcriptomic approach to the study of stage differentiation in Leishmania infantum. Proteomics 6, 3567–3581 [DOI] [PubMed] [Google Scholar]
- 48.Nie, L., Wu, G., and Zhang, W. (2006) Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics 174, 2229–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shilov, I. V., Seymour, S. L., Patel, A. A., Loboda, A., Tang, W. H., Keating, S. P., Hunter, C. L., Nuwaysir, L. M., and Schaeffer, D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.