Figure 5.
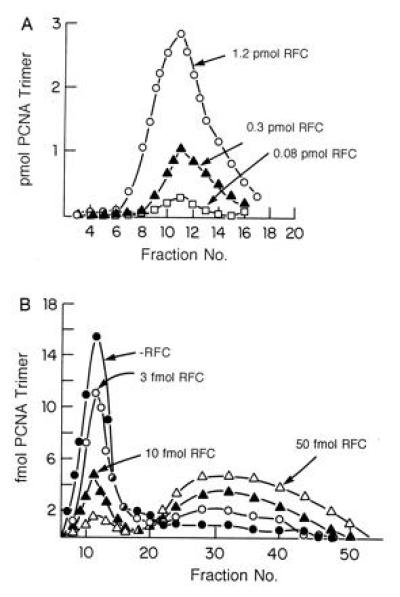
bRFC loads and unloads PCNA onto and off DNA. (A) bRFC catalyzed loading of PCNA onto singly nicked pBluescript DNA. Reaction mixtures (100 μl) containing 40 mM Tris·HCl (pH 7.5), 0.5 mM DTT, 20 μg/ml BSA, 7 mM Mg (OAc)2, 2 mM ATP, 0.5 pmol of singly nicked pBluescript plasmid DNA, 5.2 pmol of 32P-labeled PCNA trimer (574 cpm/fmol) and bRFC (as indicated), were incubated for 10 min at 37°C. Each reaction mixture was then filtered through a 6 ml Bio-Gel A15m column equilibrated with buffer containing 0.02 M Tris·HCl (pH 7.5), 0.1 mM EDTA, 40 μg/ml bovine serum albumin, 8 mM MgCl2, 4% glycerol, 5 mM DTT, and 0.1 M NaCl at 4°C. Fractions (180 μl) were collected and subjected to Cerenkov counting to detect the elution of the labeled PCNA. The peak shown in the graph represented PCNA that eluted in the excluded volume, i.e., PCNA complexed with DNA. (B) bRFC unloads 32P-PCNA catalytically from singly nicked DNA. 32P-PCNA was assembled onto DNA as described in A and the product isolated by BioGel A15m separation. The isolated PCNA–DNA complexes were then incubated in the presence or absence of bRFC (amounts as indicated) in reaction mixtures as described in A prior to gel filtration to resolve free 32P-PCNA from the 32P-PCNA–DNA complex. Both included and excluded material were subjected to Cerenkov counting as described above.
