Fig. 5.
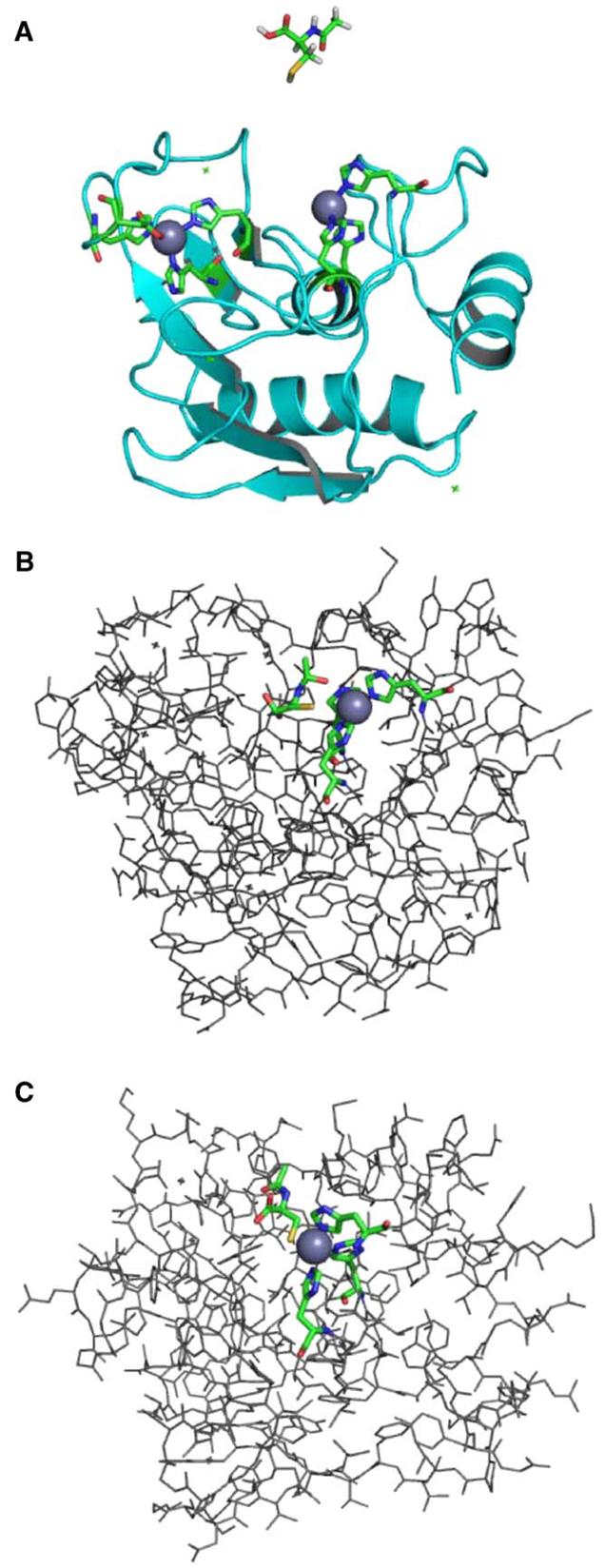
Docking of the MMP-9/NAC complex. (A) MMP-9 protein structure showing both zinc sites. The active-site zinc (on right, gray sphere), within the potential docking cleft, is coordinated by three histidines represented as sticks with the following atom coloring: carbon—green, nitrogen—blue, oxygen—red. Hydrogen atoms are not shown in the protein structure. The NAC molecule used in the docking study is shown above the MMP-9 structure. The NAC geometry was optimized at the B3LYP/6-31 + G* level of theory using Gaussian03 software [29]. The atom coloring is as above with the following additions: sulfur—yellow, hydrogen—white. (B) A docked structure of the MMP-9/NAC complex showing NAC positioned in the docking cleft nearest the active-site zinc, with the NAC sulfur within coordination distance of the metal. This structure was determined using the computational docking procedure described in the text. Hydrogen atoms are not shown. Atom coloring is as described above. (C) A model of the MMP-9/NAC complex in which the NAC ligand is coordinated to the active-site zinc analogous to Cys 99 in the crystal structure of pro-MMP-9 taken from the Protein Data Bank (Accession No. 1L6J) [45]. A partial geometry optimization was performed using the ONIOM method [46] as implemented in Gaussian03 [29] at the RHF/LANL2MB:UFF level of theory. The NAC ligand, zinc atom, and three coordinated histidines were included in the quantum mechanical region (RHF/LANL2MB), and the remaining portion of the MMP-9 protein was treated at the molecular mechanics level (UFF). Atom coloring is as described above. All graphical representations were generated with PyMol [47].
