Figure 1.
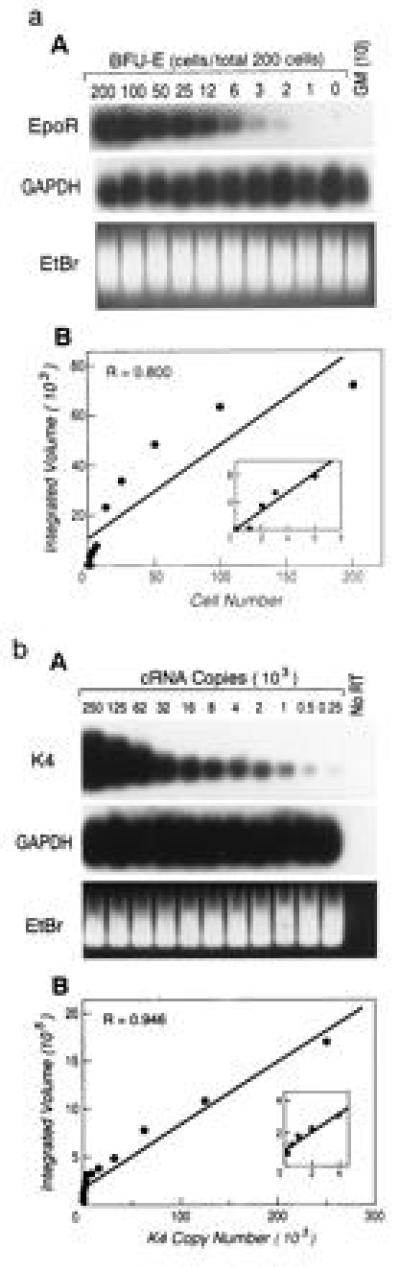
(a) Titration of input cell dose confirms the semi-quantitative nature of the modified RT-PCR technique. (A) The indicated number of cells extracted from a maturing BFU-E were diluted into cells extracted from a cell line (NIH 3T3) maintaining a constant total number of cells at 200. RT-PCR was performed as described and the resultant cDNA visualized by ethidium bromide (EtBr) staining prior to Southern transfer and hybridization with radiolabeled probes from the cDNAs from either the housekeeping gene GAPDH or the BFU-E-specific Epo-R gene. PhosphorImager quantitation of the Epo-R signal was plotted (B), and the indicated linear coefficient was determined. (Inset) Lower cell number values from the same line plot. Lanes in which 10 cells from a developing CFU-GM instead of BFU-E were tested by the RT-PCR reaction are designated GM(10). (b) Limiting dilution of known copy numbers of RNA to define the sensitivity and quantitative capacity of the single-cell RT-PCR technique. (A) An in vitro transcribed, mutated HIV-1 gag fragment, K4, was polyadenylylated, quantitated by spectrophotometry, and added in 2-fold dilutions as indicated to lysates from single CMK cells or (in independent experiments) primary quiescent bone marrow mononuclear cells prior to RT-PCR. The cDNA resulting from the RT-PCR technique was visualized following EtBr staining prior to hybridization with radiolabeled probes from the cDNAs from either GAPDH or K4 (a). PhosphorImager quantitation of the K4 signal was plotted (B) and the indicated linear coefficient was determined. (Inset) Lower K4 copy number values from the same line plot. Copy number values below 250/cell yielded a signal that was detectable but no longer linearly related to input. Lanes in which reverse transcriptase was not added to the RT-PCR mixture are designated No RT.
