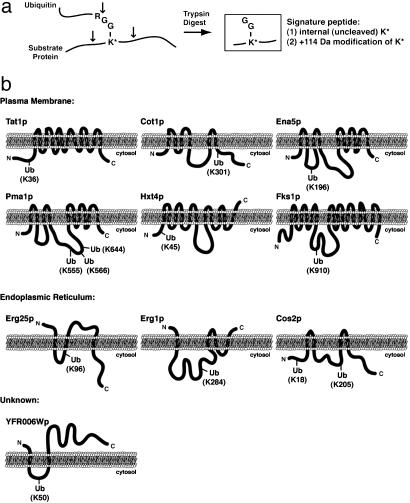
Fig. 3.
Identification of precise sites of substrate protein ubiquitination. (a) Schematic diagram indicating the ability to detect ubiquitinated lysine residues by tandem mass spectrometry. After trypsin digestion, a diglycine ubiquitin remnant remains covalently attached by an isopeptide linkage to the ε-amino side chain of a lysine residue within the substrate protein. The modified lysine, which is resistant to trypsin cleavage, can be identified by tandem mass spectrometry as a 114-Da-modified internal lysine residue. (b) Schematic representations of the predicted membrane topology and identified site(s) of ubiquitination for 15 integral membrane proteins localized to the plasma membrane, ER, and unknown membranes as indicated.