Figure 4.
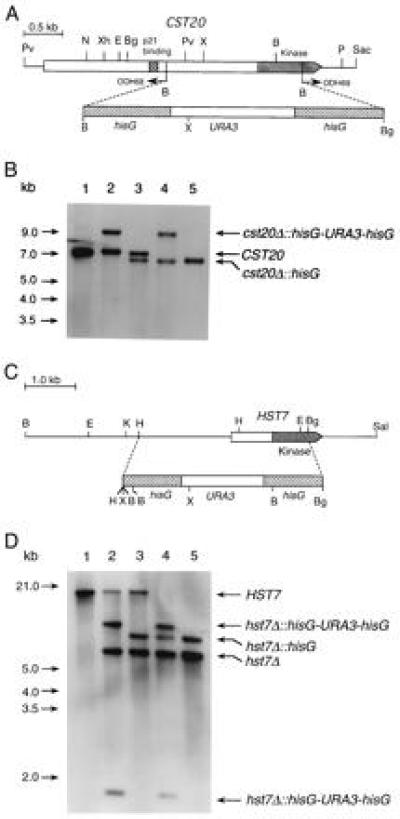
(A) Deletion of CST20 in C. albicans. PCR with the divergent oligodeoxynucleotides ODH68 and ODH69 was used to partially delete the coding sequence of CST20. A hisG-URA3-hisG cassette was then inserted. (B) Southern blot analysis with a CST20 fragment from EcoRI–XbaI as a probe. The genomic DNA samples digested with XhoI were from following strains: Lanes: 1, CAI4 (ura3/ura3 CST20/CST20); 2, CDH15 (ura3/ura3 CST20/cst20Δ::hisG-URA3-hisG); 3, CDH18 (ura3/ura3 CST20/cst20Δ::hisG); 4, CDH22 (ura3/ura3 cst20Δ::hisG-URA3hisG/cst20Δ::hisG); and 5, CDH25 (ura3/ura3 cst20Δ::hisG/cst20Δ::hisG). (C) Deletion of HST7 in C. albicans. A HindIII–BglII fragment was replaced by the hisG-URA3-hisG cassette to create plasmid pPF-7. (D) Southern blot analysis was performed with the two large BamHI fragments of pPF-7 as probes recognizing the HST7 sequence upstream of the HindIII site and the hisG-URA3-hisG cassette. The genomic DNA samples digested with XbaI were from following strains. Lanes: 1, CAI4 (ura3/ura3 HST7/HST7); 2, CDH5 (ura3/ura3 HST7/hst7Δ::hisG-URA3-hisG); 3, CDH8 (ura3/ura3 HST7/hst7Δ::hisG); 4, CDH10 (ura3/ura3 hst7Δ::hisG-URA3-hisG/hst7Δ::hisG); and 5, CDH12 (ura3/ura3 hst7Δ::hisG/hst7Δ::hisG). The endonuclease restriction sites are as follows: B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; K, KpnI; N, NsiI; P, PstI; Pv, PvuII; Sac, SacI; Sal, SalI; X, XbaI; Xh, XhoI.
