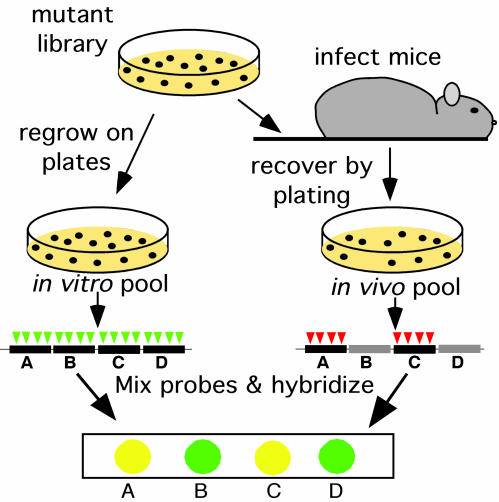
Fig. 1.
TraSH to identify attenuated mutants. C57BL/6J mice were infected intravenously with 106 colony-forming units of the mutant library. At the indicated times after infection, surviving bacteria were recovered by plating on agar medium (in vivo pools). The in vitro pool was generated by replating the library. The in vitro pool contained mutants with insertions (represented by triangles) in each nonessential gene (black bars). Mutants harboring insertions in genes that are specifically required for survival in the mouse spleen (gray bars) were lost from the in vivo pool. Genomic DNA was isolated from each pool, and TraSH probe was generated that was complementary to the chromosomal sequence flanking each insertion in the pool (4). The probes from the two pools were labeled with different colored fluorophores and mixed. Probes were then hybridized to a microarray onto which DNA fragments (features) were immobilized that were complementary to each gene in the genome of M. tuberculosis. Features that hybridized to the probe from the in vitro, but not the in vivo, pool represented genes that were specifically required for growth in the mouse.