Figure 3.
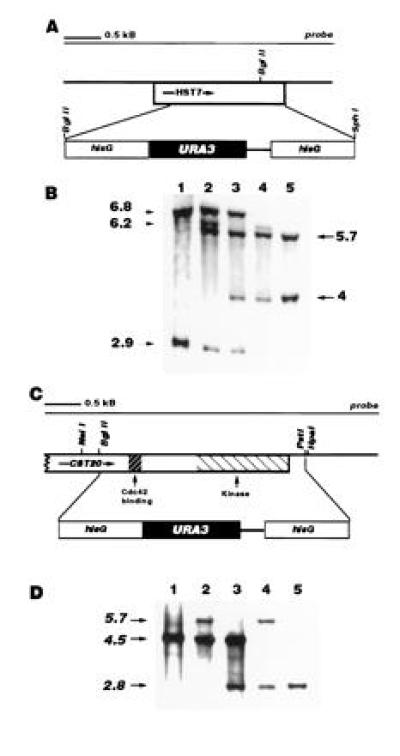
(A) HST7 deletion construct: map of pJK41. The BglII site, with which the hisG-URA3-hisG cassette was inserted, and the SphI site were created by site-directed mutagenesis and therefore not present in the wild-type gene. (B) Southern blot of HST7 deletion mutants: Candida genomic DNA digested with BglII. Lane 1, wild type. The 6.8-kb band results from digestion at the BglII site in the HST7 open reading frame and a site on the chromosome upstream of the gene, the 2.9-kb band results from digestion the BglII site in the open reading frame and a site on the chromosome downstream of it. Lane 2, HST7/hst7::hisG-URA3-hisG. Integration of the disruption construct at one of the two copies of HST7 results in loss of the BglII site in the open reading frame and gain of a new site in the construct, creating a 6.2-kb band between the new BglII site and the 3′ chromosomal BglII site, and a 5.7-kb band between the 5′ chromosomal site and the new BglII site. Lane 3, HST7/hst7::hisG. Eviction of URA3 and one hisG repeat results in a decrease in size of the fragment between the new BglII site and the 3′ site from 6.2 to 4 kb. Lane 4, hst7::hisG-URA3-hisG/hst7::hisG. Integration of the disruption construct into the remaining HST7 allele results in loss of the wild-type bands. Lane 5, hst7::hisG/hst7::hisG. Arrows indicate kilobases. Fragment sizes are approximate. (C) CST20 deletion construct: map of pJK51. The construct lacks the first 0.3 kb of the open reading frame. (D) Southern blot of CST20 mutants: Candida genomic DNA digested with NsiI. Lane 1, wild type. The band results from digestion at the NsiI site in the open reading frame and a chromosomal NsiI site 4.5 kb downstream of it. Lane 2. CST20/cst20::hisG-URA3-hisG. Lane 3, CST20/cst20::hisG. Lane 4, cst20::hisG-URA3-hisG/cst20::hisG. Lane 5, cst20::hisG/cst20::hisG. Arrows indicate kilobases. Fragment sizes are approximate.
